Vol 69 No 1 (2024): Journal of the Chilean Chemical
Short Communications
Computational Design and Toxicity Prediction of Oxazole Derivatives Targeting PPARγ as Potential Therapeutics for Diabetes Mellitus in Compare to Rosiglitazone and Pioglitazone

Published
October 6, 2024
Keywords
- PPARγ, Oxazole, Binding Site, Computer Aided Design, Drug Discovery, Diabetes Mellitus
How to Cite
Rashid, M. R., Husain, A., Mohammad Ajmal, Mausin Khan, & Sana Hashmi. (2024). Computational Design and Toxicity Prediction of Oxazole Derivatives Targeting PPARγ as Potential Therapeutics for Diabetes Mellitus in Compare to Rosiglitazone and Pioglitazone. Journal of the Chilean Chemical Society, 69(1), 6056-6064. Retrieved from https://jcchems.com/index.php/JCCHEMS/article/view/2696
Copyright (c) 2024 SChQ

This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License.
Abstract
The goal of this research is to investigate new oxazole derivative from designed series (A1-7; B1-8 & C1-8) in order to find new drug molecules for treatment of Diabetes Mellitus (DM). The PPAR receptor was chosen as the target of molecular docking investigations, which were executed using PyRx software. In silico analyses, including physicochemical properties, drug score, drug likeness, solubility, and toxicity prediction, were conducted using software such as Swiss ADME, Osiris property explorer, Lipinski filter and Toxtree method. All molecules passed the Lipinski rule with the zero violations and synthetic score was also found to be in the easy limit. All ligands showed drug score values ranging from 0.11 to 0.9 (no negative value). Compounds A6, C2, C5, C6, C7 and C8 were shown drug score from 0.91 to 0.80, which is closer to 1 and therefore considered as druggable ligands, when compared with the standard drug, Rosiglitazone and Pioglitazone also found non-toxic. All compounds shown logP values between -0.25 to 4.58. The RMSD value of receptor and receptor-ligand complexes was analyzed, and it revealed the stability of binding interactions and remained stable throughout the simulation. Compound C8 was found highest RMSD score (67.34Å) in compare to other compounds and standard drug Rosiglitazone (64.31Å). The TPSA were found within the range 35.26 to 128.60 and MR also were in the range 32.21-113.62. Compounds were found to be non-substrate for p glycoprotein except C4, high GIA% (>90%), also displayed negative permeability across the BBB, and most of compounds were found inhibitor of CYP 1A2 and CYP 2C19 and non-inhibitor of CYP 2C9, CYP 2D6 and CYP 3A4. Compound C5 was exhibited higher drug score (0.91), bioactivity score and revealed good drug relevant properties, ADME and no toxicity profile in compared to other ligands and standard drugs. The most active compound of the series was found C5 and C8 therefore further studies on this compound continue in our research laboratory to acquire more information about SAR and QSAR. Finally, it is conceivable that further derivatization of these compounds could result in obtaining more selective lead compounds.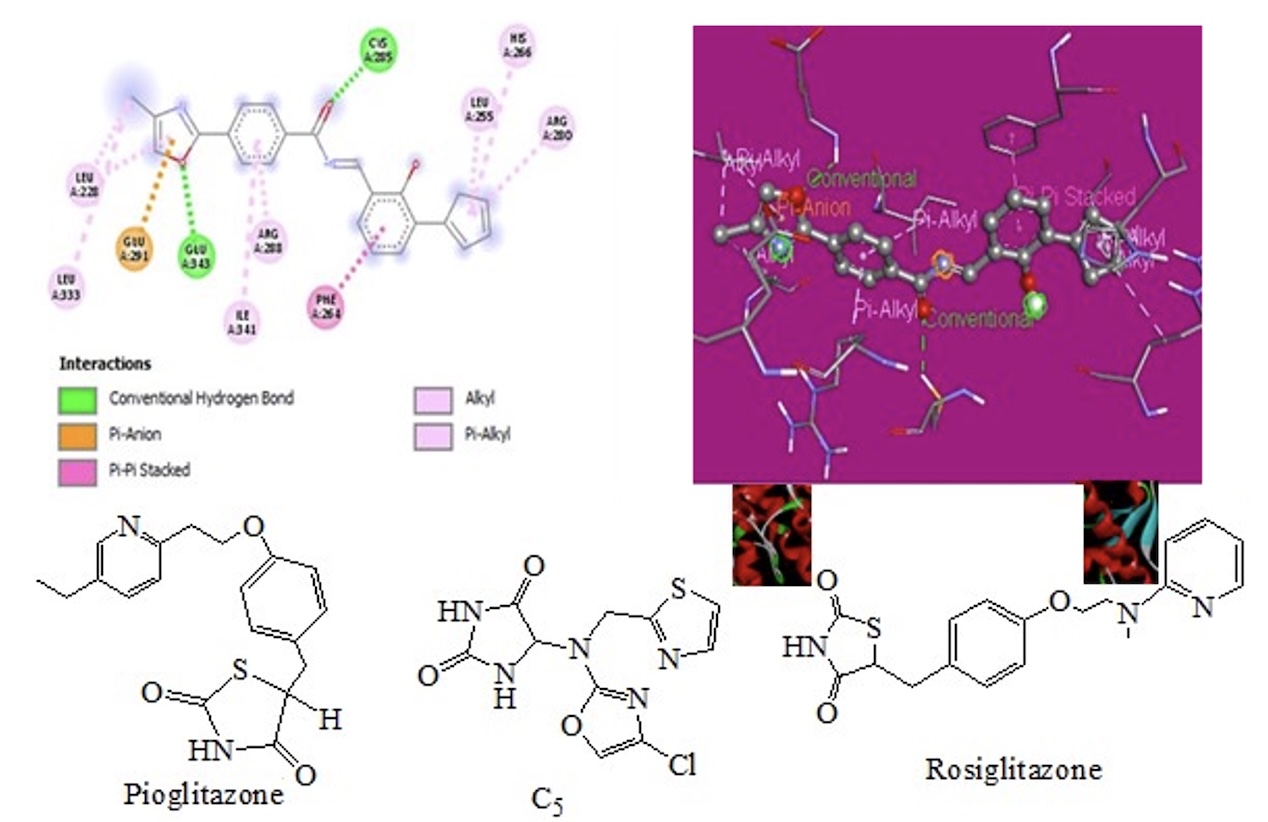
References
- References
- Rani A, Arora S, Goyal A. Antidiabetic plants in traditional medicines: A review. International Research Journal of Pharmacy. 2017; 8: 17-24.
- Deshmukh CD, Jain A, Nahata B. Diabetes mellitus: a review. International Journal of Pure and Applied Biosciences. 2015; 3: 224-30.
- Adapa D, Sarangi TK. A review on diabetes mellitus: complications, management and treatment modalities. Journal of Medical Health and Sciences. 2015; 4(3): 331-39.
- Okur ME, Karantas ID, Siafaka PI. Diabetes Mellitus: a review on pathophysiology, current status of oral pathophysiology, current status of oral medications and future perspectives. ACTA Pharmaceutica Science. 2017; 55(1): 231-42.
- Chaudhury A, Duvoor C, Reddy Dendi VS, Kraleti S, Chada A, Ravilla R, Marco A, Shekhawat NS, Montales MT, Kuriakose K, Sasapu A, Beebe A, Patil N, Musham CK, Lohani GP, Mirza W. Clinical review of antidiabetic drugs: implications for type 2 diabetes mellitus management. Front Endocrinol (Lausanne). 2017; 8: 6. doi: 10.3389/fendo.2017.00006. PMID: 28167928.
- Yadav M, Saraswat N, Wal P, Rai A, Singh D. A comparative study of drug interaction and side effect of drug for treatment of Diabetes Mellitus: A Review. International Research Journal of Pharmacy. 2018; 9(6): 14-6.
- Inzucchi SE. Oral antihyperglycemic therapy for type 2 diabetes: scientific review. JAMA. 2002; 287(3): 360-72.
- Shah N, Abdalla MA, Deshmukh H, Sathyapalan T. Therapeutics for type-2 diabetes mellitus: a glance at the recent inclusions and novel agents under development for use in clinical practice. Ther Adv Endocrinol Metab. 2021 Sep 23; 12: 20420188211042145. doi: 10.1177/20420188211042145. PMID: 34589201.
- Nguyen ND, LeL T. Targeted proteins for diabetes drug design. Advances in Natural Sciences: Nanoscience and Nanotechnology. 2012; 3(1): 013001.
- Qaoud MT, Almasri I, Önkol T. Peroxisome proliferator-activated receptors as superior targets for treating diabetic disease, design strategies - review article. Turk J Pharm Sci. 2022; 19(3): 353-370. doi: 10.4274/tjps.galenos.2021.70105. PMID: 35775494.
- Holm LJ, Mønsted MØ, Haupt-Jorgensen M, Buschard K. PPARs and the Development of Type 1 Diabetes. PPAR Res. 2020 Jan 9; 2020: 6198628. doi: 10.1155/2020/6198628. PMID: 32395123.
- Kume S, Uzu T, Isshiki K, Koya D. Peroxisome proliferator-activated receptors in diabetic nephropathy. PPAR Res. 2008; 2008: 879523. doi: 10.1155/2008/879523. PMID: 19277201.
- Zhang HZ, Zhao ZL, Zhou CH. Recent advance in oxazole-based medicinal chemistry. European journal of medicinal chemistry. 2018; 144: 444-92.
- Swellmeen L. 1, 3-Oxazole derivatives: a review of biological activities as antipathogenic. Der Pharma Chemica. 2016; 8(13): 269-86.
- Chaudhary KK, Mishra N. A review on molecular docking: novel tool for drug discovery. Databases. 2016; 3(4): 1029.
- Mani S, Supriya T, Shankar M, Kavya Lalitha S, Dastgiri J, Niranjan Babu M. An overview on molecular docking. American journal of biological and pharmaceutical research.2016; 3(2): 83-9.
- Daina A, Michielin O, Zoete V. Swiss ADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Scientific reports. 2017; 7(1): 1-3.
- Das PS, Saha P, Abdul AP. A review on computer aided drug design in drug discovery. World Journal of Pharmacy and Pharmaceutical Sciences. 2017; 6(7): 279-91.
- Hoque I, Chatterjee A, Bhattacharya S, Biswas R. An approach of computer-aided drug design (CADD) tools for in-silico pharmaceutical drug design and development. International Journal of Advances Research in Biological Sciences. 2017; 4(2): 60-71.
- Barrawaz AY, Baig SS, Salam BA. Computer aided drug design: a mini review. Journal of medical pharmaceutical and allied sciences. 2020; 1(5): 2584-2591.
- GonzáLez-Serrano JI, Sánchez-Portal M, Castañeda H, Quirk R, De Miguel ED, Aguiar M, Cepa J. OSIRIS software: The Mask designer tool. Experimental Astronomy. 2004; 18(1): 65-75.
- Basuki SA, Maulanasari NM, Astuti EJ. Toxicity on class of antibiotic agents using Toxtree software and its interaction with its receptors using molecular virtual docker software. In Health Science International Conference (HSIC 2017) 2017; 175-180. DOI:10.2991/hsic-17.2017.28
- Ravi L, Kannabiran K. A Handbook on protein-ligand docking tool: Auto Dock 4. Innovare Journal of Medical Sciences. 2016; 3: 28-33.
- Morris GM, Goodsell DS, Pique ME, Lindstrom WL, Huey R, Forli S, Hart WE, Halliday S, Belew R, Olson AJ. Automated docking of flexible ligands to flexible receptors. User guide Auto Dock. 2010.
- Dundas J, Ouyang Z, Tseng J, Binkowski A, Turpaz Y, Liang J. CASTp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic acids research. 2006; 34(2): 116-8.
- Walters WP. Going further than Lipinski's rule in drug design. Expert Opinion on Drug Discovery. 2012; 7(2): 99-107.
- Daina A, Zoete V. Application of the Swiss drug design on line resources in virtual screening. International journal of molecular sciences. 2019; 20(18): 4612.
- Ranjith D, Ravikumar C. Swiss ADME predictions of pharmacokinetics and drug-likeness properties of small molecules present in Ipomoea mauritiana Jacq. Journal of Pharmacognosy and Phytochemistry. 2019; 8(5): 2063-73.
- Rajalakshmi R, Lalitha P, Sharma SC, Rajiv A, Chithambharan A, Ponnusamy A. In Silico studies: Physicochemical properties, drug score, toxicity predictions and molecular docking of organo sulphur compounds against Diabetes mellitus. Journal of Molecular Recognition. 2021; 34(11): e2925.
- Jamkhande PG, Ghante MH, Ajgunde BR. Software based approaches for drug designing and development: A systematic review on commonly used software and its applications. Bulletin of Faculty of Pharmacy, Cairo University. 2017; 55(2): 203-10.
- Huey R, Morris GM. Using Auto Dock4 with Auto Dock tools: a tutorial. The Scripps Research Institute, USA. 2008; 8: 54-6.
- B Fernandes T, CF Segretti M, C Polli M, Parise-Filho R. Analysis of the applicability and use of Lipinskis rule for central nervous system drugs. Letters in Drug Design & Discovery. 2016; 13(10): 999-1006.
- Brito MA. Pharmacokinetic study with computational tools in the medicinal chemistry course. Brazilian Journal of Pharmaceutical Sciences. 2011; 47: 797-805.
- Doak BC, Over B, Giordanetto F, Kihlberg J. Oral druggable space beyond the rule of 5: insights from drugs and clinical candidates. Chemistry & biology. 2014; 21(9): 1115-42.
- Manikandan P, Nagini S. Cytochrome P450 structure, function and clinical significance: are view. Current drug targets. 2018; 19(1): 38-54.
- Srivastava R. The critical studies on the molecular properties, toxicity, and biological efficacy of 21 new chemical entities. ACS omega. 2021; 6(38): 24891-901.
- Rashid M, Md Tanwir A, Mohammed Abdelmageed, Mohammed Hilal M Al‑Harbi, Asif Husain, Dheeraj Bisht, Rajeshwar Kamal Kant Arya. Silver nanoparticles from Saudi and Syrian black cumin seed extracts: green synthesis, ADME, toxicity, comparative research, and biological appraisal, Journal of Pharmacy and Bioallied Sciences. 2023, 15 (4), 52-64.
- Bharath EN, Manjula SN, Vijaych. In silico drug design tool for overcoming the innovation deficit in the drug discovery process. International Journal of Pharmacy and Pharmaceutical Sciences. 2011; 3(2): 8-12.
- Rashid Mohammad, Obaid Afzal and Abdul Malik Saleh Alfawaz Altamimi, Benzimidazole molecules hybrid with oxadiazole ring as antiproliferative agent: In-Silico Analysis, Synthesis and Biological Evaluation, Journal of Chilean Chemical Society, 2021; 66 (2): 5164-5182.
- Sachin MM, Sachin MM, Manoj SG, Mandar AT, Rinkesh MT. A Review: In-Silico approaches in predictive toxicology. International Journal of Pharmaceutical Sciences Review and Research. 2021; 70(2): 100-108.