- Dengue Virus, Envelope Glycoprotein, Molecular Docking, ADMET, Molecular Dynamic
Copyright (c) 2024 SChQ

This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License.
Abstract
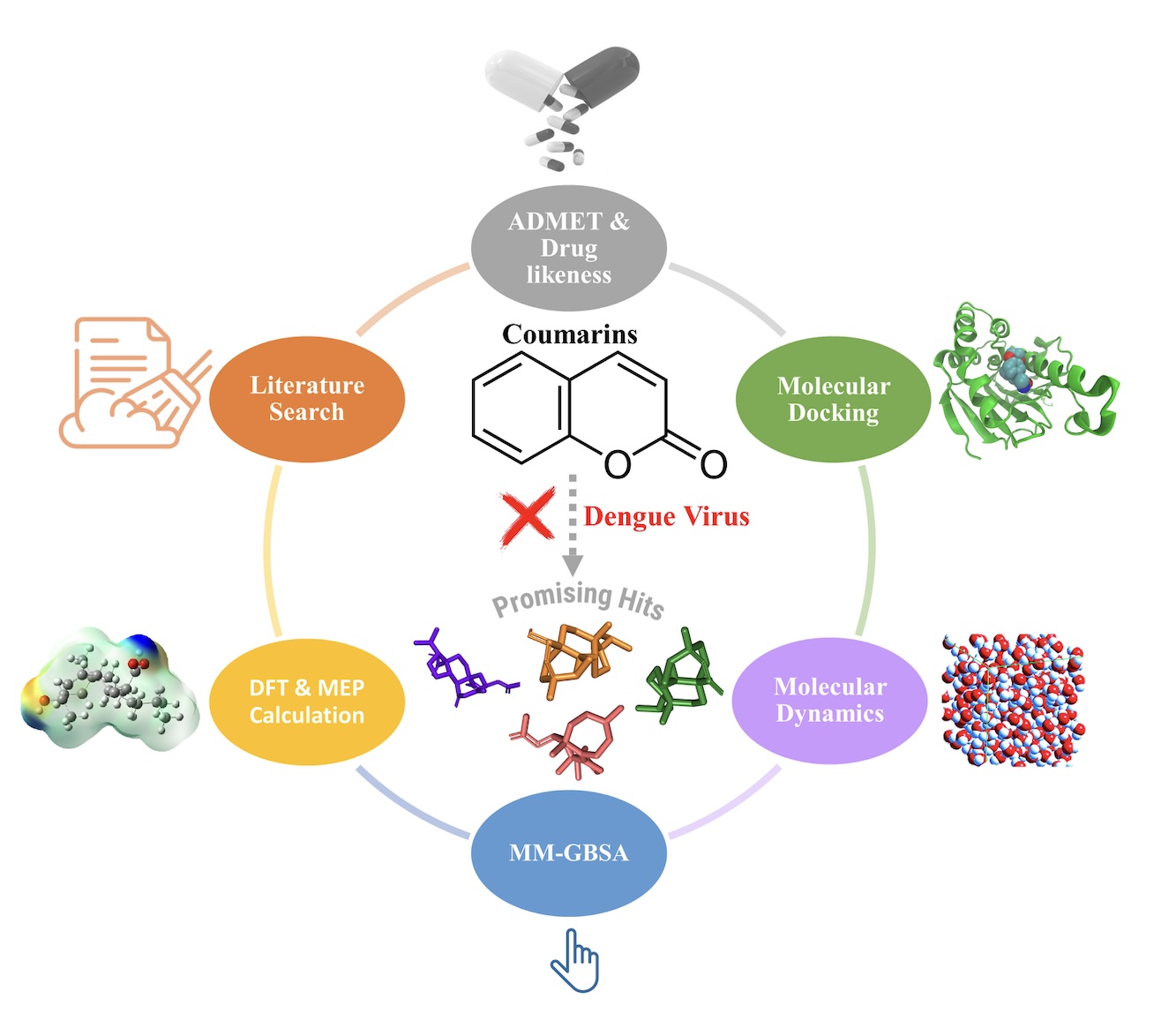
Dengue is a virus-borne disease that causes fever, headaches, nausea, muscle pain, and other symptoms. The majority of cases are mild, but there are severe forms of Dengue fever like hemorrhagic fever and Dengue shock syndrome, which can be life-threatening. Severe cases may cause breathing difficulties, excessive bleeding, abdominal pain, vomiting, fluid build-up, and extreme fatigue. This highlights the need for widespread knowledge and health care interventions in areas where Dengue is prevalent. . The objective of this work was to use virtual screening to explore how coumarin derivatives bind to Dengue virus protein targets and their affinities to ligands and receptors. Molegro Virtual Docker allowed for structure based virtual screening by taking advantage of the importance of protein like interactions in drug development. The docking studies provide a basis to understand the early stages of ligand-receptor interactions and guide further research. In order to further investigate the complex atomic-level behaviours of proteins and biomolecules in the fields of molecular biology and drug discovery, we performed estimations using molecular dynamics simulations (MD) and MM-GBSA calculations. The evaluation encompassed the examination of antiviral activity, drug-likeness, ADMET features, and quantum chemistry techniques of selected compounds. This comprehensive analysis aimed to explore the molecular interactions between coumarin derivatives and Dengue virus protein targets from multiple perspectives. The findings indicate that (S)-6-hydroxy-7-(5-hydroxy-3,7-dimethyl-2,6-octadienyloxy)coumarin demonstrates the greatest binding affinity among the chemicals that were examined. It is then followed by 6-hydroxy-7-(7-hydroxy-3,7-dimethyl-2,5-octadienyloxy)coumarin, wedelolactone, and medicagol. These findings have the potential to enhance the progress of creating new antiviral drugs that are more effective in treating Dengue virus infections, by utilising various artificial intelligence methods.
References
- O. J. Brady, P. W. Gething, S. Bhatt, J. P. Messina, J. S. Brownstein, A. G. Hoen, C. L. Moyes, A. W. Farlow, T. W. Scott, and S. I. Hay, (2012).
- J. Whitehorn and C. P. Simmons, Vaccine 29, 7221 (2011).
- S. Mishra, A. Pandey, and S. Manvati, Heliyon 6, (2020).
- S. Bhatt, P. W. Gething, O. J. Brady, J. P. Messina, A. W. Farlow, C. L. Moyes, J. M. Drake, J. S. Brownstein, A. G. Hoen, and O. Sankoh, Nature 496, 504 (2013).
- J. D. Stanaway, D. S. Shepard, E. A. Undurraga, Y. A. Halasa, L. E. Coffeng, O. J. Brady, S. I. Hay, N. Bedi, I. M. Bensenor, and C. A. Castañeda-Orjuela, Lancet Infect. Dis. 16, 712 (2016).
- S. R. Mutheneni, A. P. Morse, C. Caminade, and S. M. Upadhyayula, Emerg. Microbes Infect. 6, 1 (2017).
- F. Meng, R. A. Badierah, H. A. Almehdar, E. M. Redwan, L. Kurgan, and V. N. Uversky, FEBS J. 282, 3368 (2015).
- V. D. Dwivedi, I. P. Tripathi, R. C. Tripathi, S. Bharadwaj, and S. K. Mishra, Brief. Funct. Genomics 16, 217 (2017).
- L. R. Souza, J. G. Colonna, J. M. Comodaro, and F. G. Naveca, BMC Bioinformatics 23, 1 (2022).
- H. K. H. Luk, X. Li, J. Fung, S. K. P. Lau, and P. C. Y. Woo, Infect. Genet. Evol. 71, 21 (2019).
- P. Bhatt, S. P. Sabeena, M. Varma, and G. Arunkumar, Curr. Microbiol. 78, 17 (2021).
- H. A. Imad, W. Phumratanaprapin, B. Phonrat, K. Chotivanich, P. Charunwatthana, S. Muangnoicharoen, S. Khusmith, T. Tantawichien, J. Phadungsombat, and E. Nakayama, Am. J. Trop. Med. Hyg. 102, 943 (2020).
- X. Pang, R. Zhang, and G. Cheng, Virol. Sin. 32, 16 (2017).
- K. N. Venugopala, V. Rashmi, and B. Odhav, Biomed Res. Int. 2013, (2013).
- A. Stefanachi, F. Leonetti, L. Pisani, M. Catto, and A. Carotti, Molecules 23, 250 (2018).
- S. D. Sarker and L. Nahar, Prog. Chem. Org. Nat. Prod. 106 241 (2017).
- L. P. Jigar, The Introduction of Coumarin (Blue Rose Publishers, 2019).
- N. H. Jadhav, S. S. Sakate, N. K. Rasal, D. R. Shinde, and R. A. Pawar, ACS Omega 4, 8522 (2019).
- M. Z. Hassan, H. Osman, M. A. Ali, and M. J. Ahsan, Eur. J. Med. Chem. 123, 236 (2016).
- R. W. DeSimone, K. S. Currie, S. A. Mitchell, J. W. Darrow, and D. A. Pippin, Comb. Chem. High Throughput Screen. 7, 473 (2004).
- D. Mathew and W.-L. Hsu, J. Funct. Foods 40, 692 (2018).
- J. Zhu and J. Jiang, Mol. Nutr. Food Res. 62, 1701073 (2018).
- U. Laila, M. Akram, M. A. Shariati, A. M. Hashmi, N. Akhtar, I. M. Tahir, A. O. Ghauri, N. Munir, M. Riaz, and N. Akhter, Clin. Exp. Pharmacol. Physiol. 46, 1063 (2019).
- D. Srikrishna, C. Godugu, and P. K. Dubey, Mini Rev. Med. Chem. 18, 113 (2018).
- E. Kudo, M. Taura, K. Matsuda, M. Shimamoto, R. Kariya, H. Goto, S. Hattori, S. Kimura, and S. Okada, Bioorg. Med. Chem. Lett. 23, 606 (2013).
- B. T. Dharmapalan, R. Biswas, S. Sankaran, B. Venkidasamy, M. Thiruvengadam, G. George, M. Rebezov, G. Zengin, M. Gallo, and D. Montesano, Viruses 14, 2656 (2022).
- G. M. Morris, R. Huey, W. Lindstrom, M. F. Sanner, R. K. Belew, D. S. Goodsell, and A. J. Olson, J. Comput. Chem. 30, 2785 (2009).
- D. B. Kitchen, H. Decornez, J. R. Furr, and J. Bajorath, Nat. Rev. Drug Discov. 3, 935 (2004).
- R. Thomsen and M. H. M. CHRISTENSEN, J. Med. Chem. 3315 (n.d.).
- T. Xu, A. Sampath, A. Chao, D. Wen, M. Nanao, P. Chene, S. G. Vasudevan, and J. Lescar, J. Virol. 79, 10278 (2005).
- D. Luo, T. Xu, R. P. Watson, D. Scherer-Becker, A. Sampath, W. Jahnke, S. S. Yeong, C. H. Wang, S. P. Lim, A. Strongin, S. G. Vasudevan, and J. Lescar, EMBO J. 27, 3209 (2008).
- D. Luo, N. Wei, D. N. Doan, P. N. Paradkar, Y. Chong, A. D. Davidson, M. Kotaka, J. Lescar, and S. G. Vasudevan, J. Biol. Chem. 285, 18817 (2010).
- C. M. D. Swarbrick, C. Basavannacharya, K. W. K. Chan, S.-A. Chan, D. Singh, N. Wei, W. W. Phoo, D. Luo, J. Lescar, and S. G. Vasudevan, Nucleic Acids Res. 45, 12904 (2017).
- S. P. Lim, L. S. Sonntag, C. Noble, S. H. Nilar, R. H. Ng, G. Zou, P. Monaghan, K. Y. Chung, H. Dong, B. Liu, C. Bodenreider, G. Lee, M. Ding, W. L. Chan, G. Wang, Y. L. Jian, A. T. Chao, J. Lescar, Z. Yin, T. R. Vedananda, T. H. Keller, and P.-Y. Shi, J. Biol. Chem. 286, 6233 (2011).
- B. Coutard, E. Decroly, C. Li, A. Sharff, J. Lescar, G. Bricogne, and K. Barral, Antiviral Res. 106, 61 (2014).
- M. B. Brecher, Z. Li, J. Zhang, H. Chen, Q. Lin, B. Liu, and H. Li, Protein Sci. 24, 117 (2015).
- C. G. Noble, S.-H. Li, H. Dong, S. H. Chew, and P.-Y. Shi, Antiviral Res. 111, 78 (2014).
- M. Brecher, H. Chen, Z. Li, N. K. Banavali, S. A. Jones, J. Zhang, L. D. Kramer, and H. Li, ACS Infect. Dis. 1, 340 (2015).
- F. Benmansour, I. Trist, B. Coutard, E. Decroly, G. Querat, A. Brancale, and K. Barral, Eur. J. Med. Chem. 125, 865 (2017).
- M. Feracci, C. Eydoux, V. Fattorini, L. Lo Bello, P. Gauffre, B. Selisko, P. Sutto-Ortiz, A. Shannon, H. Xia, P.-Y. Shi, M. Noel, F. Debart, J.-J. Vasseur, S. Good, K. Lin, A. Moussa, J.-P. Sommadossi, A. Chazot, K. Alvarez, J.-C. Guillemot, E. Decroly, F. Ferron, and B. Canard, Antiviral Res. 212, 105574 (2023).
- S. P. Lim, C. G. Noble, C. C. Seh, T. S. Soh, A. El Sahili, G. K. Y. Chan, J. Lescar, R. Arora, T. Benson, S. Nilar, U. Manjunatha, K. F. Wan, H. Dong, X. Xie, P.-Y. Shi, and F. Yokokawa, PLoS Pathog. 12, e1005737 (2016).
- C. G. Noble, S. P. Lim, R. Arora, F. Yokokawa, S. Nilar, C. C. Seh, S. K. Wright, T. E. Benson, P. W. Smith, and P.-Y. Shi, J. Biol. Chem. 291, 8541 (2016).
- H. Shimizu, A. Saito, J. Mikuni, E. E. Nakayama, H. Koyama, T. Honma, M. Shirouzu, S.-I. Sekine, and T. Shioda, PLoS Negl. Trop. Dis. 13, e0007894 (2019).
- R. Arora, C. W. Liew, T. S. Soh, D. A. Otoo, C. C. Seh, K. Yue, S. Nilar, G. Wang, F. Yokokawa, C. G. Noble, Y. L. Chen, P.-Y. Shi, J. Lescar, T. M. Smith, T. E. Benson, and S. P. Lim, J. Virol. 94, (2020).
- C. G. Noble, C. C. Seh, A. T. Chao, and P. Y. Shi, J. Virol. 86, 438 (2012).
- M. Yildiz, S. Ghosh, J. A. Bell, W. Sherman, and J. A. Hardy, ACS Chem. Biol. 8, 2744 (2013).
- Y. Modis, S. Ogata, D. Clements, and S. C. Harrison, Proc. Natl. Acad. Sci. U. S. A. 100, 6986 (2003).
- C. N Powers and W. N Setzer, Comb. Chem. High Throughput Screen. 19, 516 (2016).
- T. A. Halgren, J. Comput. Chem. 17, 490 (1996).
- Y. Pan, N. Huang, S. Cho, and A. D. Mackerell, J. Chem. Inf. Comput. Sci. 43, 267 (2003).
- J. Yang and T. Shen, Proteins Struct. Funct. Bioinforma. 59, 205 (2005).
- N. Huang, A. Nagarsekar, G. Xia, J. Hayashi, and A. D. MacKerell, J. Med. Chem. 47, 3502 (2004).
- C. N. Hancock, A. Macias, E. K. Lee, S. Y. Yu, A. D. MacKerell, and P. Shapiro, J. Med. Chem. 48, 4586 (2005).
- C. Abad-Zapatero and J. T. Metz, Drug Discov. Today 10, 464 (2005).
- G. Carta, A. J. S. Knox, and D. G. Lloyd, J. Chem. Inf. Model. 47, 1564 (2007).
- M. Snow Setzer, K. G. Byler, I. V. Ogungbe, and W. N. Setzer, Sci. Pharm. 85, 5 (2017).
- S. Release, Maest. Interoperability Tools, Schrödinger, New York, NY (2017).
- A. Daina, O. Michielin, and V. Zoete, Sci. Rep. 7, 1 (2017).
- Y. H. Zhao, M. H. Abraham, J. Le, A. Hersey, C. N. Luscombe, G. Beck, B. Sherborne, and I. Cooper, Pharm. Res. 19, 1446 (2002).
- D. E. V Pires, T. L. Blundell, and D. B. Ascher, J. Med. Chem. 58, 4066 (2015).
- C. B. Jalkute, S. H. Barage, M. J. Dhanavade, and K. D. Sonawane, Int. J. Pept. Res. Ther. 21, 107 (2015).
- Hypercube, (2007).
- M. J. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, G. Scalmani, V. Barone, B. Mennucci, G. A. Petersson, H. Nakatsuji, M. Caricato, X. Li, H. P. Hratchian, A. F. Izmaylov, J. Bloino, G. Zheng, J. L. Sonnenberg, M. Hada, M. Ehara, K. Toyota, R. Fukuda, J. Hasegawa, M. Ishida, T. Nakajima, Y. Honda, O. Kitao, H. Nakai, T. Vreven, J. A. Montgomery, J. E. Peralta, F. Ogliaro, M. Bearpark, J. J. Heyd, E. Brothers, K. N. Kudin, V. N. Staroverov, R. Kobayashi, J. Normand, K. Raghavachari, A. Rendell, J. C. Burant, S. S. Iyengar, J. Tomasi, M. Cossi, N. Rega, J. M. Millam, M. Klene, J. E. Knox, J. B. Cross, V. Bakken, C. Adamo, J. Jaramillo, R. Gomperts, R. E. Stratmann, O. Yazyev, A. J. Austin, R. Cammi, C. Pomelli, J. W. Ochterski, R. L. Martin, K. Morokuma, V. G. Zakrzewski, G. A. Voth, P. Salvador, J. J. Dannenberg, S. Dapprich, A. D. Daniels, Ö. Farkas, J. B. Foresman, J. V Ortiz, J. Cioslowski, and D. J. Fox, (2009).
- D. R. D. K. T. A. M. JM, Gaussian Inc (2008).
- H. O. Rasul, B. K. Aziz, D. D. Ghafour, and A. Kivrak, Mol. Divers. (2022).
- L. Martínez, PLoS One 10, e0119264 (2015).
- R. Alnajjar, A. Mostafa, A. Kandeil, and A. A. Al-Karmalawy, Heliyon 6, e05641 (2020).
- P. Geerlings, F. De Proft, and W. Langenaeker, Chem. Rev. 103, 1793 (2003).
- P. K. Chattaraj, Chemical Reactivity Theory : A Density Functional View, First (CRC Press/Taylor & Francis, Boca Ratón, 2009).
- R. G. Parr and W. Yang, J. Am. Chem. Soc. 106, 4049 (1984).
- F. L. Hirshfeld, Theor. Chim. Acta 44, 129 (1977).
- A. Kumar, C. G. Mohan, and P. C. Mishra, J. Mol. Struct. THEOCHEM 361, 135 (1996).