IDENTIFICATION OF NOVEL COUMARIN BASED COMPOUNDS AS POTENTIAL INHIBITORS OF THE 3-CHYMOTRYPSIN-LIKE MAIN PROTEASE OF SARS-COV-2 USING DFT, MOLECULAR DOCKING AND MOLECULAR DYNAMICS SIMULATION STUDIES
- COVID-19, SARS-CoV-2, Coumarin, Molecular docking, Molecular dynamics, Density functional theory
Copyright (c) 2022 SChQ

This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License.
Abstract
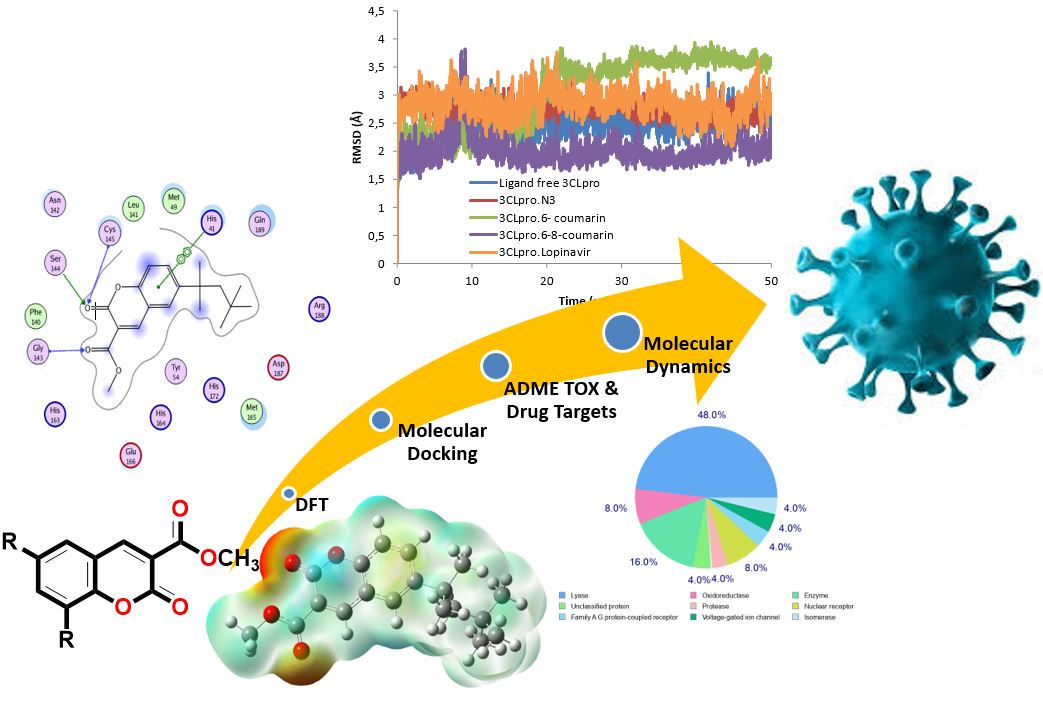
SARS-CoV-2 is the pandemic disease-causing agent COVID-19 with high infection rates. Despite the progress made in vaccine development, there is an urgent need for the identification of antiviral compounds that can tackle better the different phases of SARS-CoV-2. The main protease (Mpro or 3CLpro) of SARS-CoV-2 has a crucial role in viral replication and transcription. In this study, an in silico method was executed to elucidate the inhibitory potential of the synthesized 6-tert-octyl and 6-8-ditert-butyl coumarin compounds against the major protease of SARS-CoV-2 by comprehensive molecular docking and density functional theory (DFT), ADMET properties and molecular dynamics simulation approaches. Both compounds shown favorable interactions with the 3CLpro of the virus. From DFT calculations, HOMO-LUMO values and global descriptors indicated promising results for these compounds. Furthermore, molecular dynamics studies revealed that these ligand-receptor complexes remain stable during simulations and both compounds showed considerably high binding affinity to the main SARS-CoV-2 protease. The results of the study suggest that the coumarin compounds 6-tert-octyl and 6-8-ditert-butyl could be considered as promising scaffolds for the development of potential COVID-19 inhibitors after further studies.
References
2. AJ. Rodríguez-Morales, K. MacGregor, S. Kanagarajah, D. Patel, P. Schlagenhauf, Travel. Med. Infect. Dis. 33, 101578, (2020).
3. Z. Wang, X. Chen, Y. Lu, F. Chen, W. Zhang, Biosci. Trends. (2020).
4. O. Yañez, MI. Osorio, E. Uriarte, C. Areche, W. Tiznado, JM. Pérez-Donoso, O. García-Beltrán, F. González-Nilo, Front. Chem. 8, 1273 (2021).
5. S. Hiremath, HV. Kumar, M. Nandan, M. Mantesh, K. Shankarappa, V. Venkataravanappa, CJ. Basha, CL. Reddy, 3Biotech. 11, 1-18 (2021).
6. JR. Mora, SA. Cuesta, A. Belhassan, G. Salgado Morán, T. Lakhlifi, M. Bouachrine, C. Peña, L. Gerli, LH. Mendoza-Huízar, (2022).
7. H. Zakaryan, E. Arabyan, A. Oo, K. Zandi, Arch. Virol. 162, 2539-2551 (2017).
8. SM. Thayil, S. ThyagarajanInt, J. Pharmacogn. Phytochem. Res. 8, 1020-1024 (2016).
9. S. Jo, S. Kim, DH. Shin, M-S. Kim, J. Enzyme. Inhib. Med. Chem. 35, (1):145-151, (2020).
10. IE. Orhan, FSS. Deniz, Nat. Prod. Bioprospect. 10, 171-186 (2020).
11. JS. Mani, JB. Johnson, JC. Steel, DA. Broszczak, PM. Neilsen, KB. Walsh, M. Naiker, Virus. Res. 284, 197989 (2020).
12. AK. Maurya, N. Mishra, J. Biomol. Struct. Dyn. 39, 7306-7321, (2021).
13. SK. Chidambaram, D. Ali, S. Alarifi, S. Radhakrishnan, I. Akbar, J. Infect. Public. Health. 13, 1671-1677 (2020).
14. M. Özdemir, B. Köksoy, D. Ceyhan, M. Bulut, B. YALCİN, J. Turkish. Chem. Soc. 7, 691-712, (2020).
15. M. Özdemir, B. Köksoy, D. Ceyhan, K. Sayın, E. Erçağ, M. Bulut, B. Yalçın, J. Biomol. Struct. Dyn. 1-16, (2020)
16. UR. Abdelmohsen, A. Albohy, BS. Abdulrazik, SA. Bayoumi, LG. Malak, IS. Khallaf, G. Bringmann, SF. Farag, RSC. Advances. 11, 16970-16979, (2021).
17. S. Chidambaram, MA. El-Sheikh, AH. Alfarhan, S. Radhakrishnan, I. Akbar, Saudi. J. Biol. Sci. 28, 1100-1108, (2021).
18. R. Abdizadeh, F. Hadizadeh, T. Abdizadeh, Mol. Divers. 1-24, (2021).
19. YW. Chen, C-PB. Yiu, K-Y. Wong, F1000Research. 9, (2020).
20. TW. Shattuck, Montreal, QC: Chemical Computing Group ULC, 2011.
21. OM. Zárraga, M. Darouch, E. Lisboa, PP. Arroyo, MA. Miranda, J. Chil. Chem. Soc. 66, 5220-5222 (2021).
22. M. Frisch, G. Trucks, H. Schlegel, G. Scuseria, M. Robb, J. Cheeseman, G. Scalmani, V. Barone, B. Mennucci, G. Petersson, Gaussian 09 revision D. 01,(Gaussian Inc., 2009).
23. CA. Lipinski, F. Lombardo, BW. Dominy, PJ. Feeney, Adv. Drug. Deliv. Rev. 23, 3-25, (1997)
24. DF. Veber, SR. Johnson, H-Y. Cheng, BR. Smith, KW. Ward, KD. Kopple, J. Med. Chem. 45, 2615-2623, (2002).
25. WJ. Egan, KM. Merz, JJ. Baldwin, J. Med. Chem. 43, 3867-3877, (2000).
26. I. Muegge, SL. Heald, D. Brittelli, J. Med. Chem. 44, 1841-1846, (2001).
27. RK. Goel, D. Singh, A. Lagunin, V. Poroikov, Med. Chem. Res. 20, 1509-1514, (2011).
28. N. Khurana, MPS. Ishar, A. Gajbhiye, RK. Goel, Eur. J. Pharmacol. 662, 22-30, (2011).
29. A. Srivastava, S. Siddiqui, R. Ahmad, S. Mehrotra, B. Ahmad, A. Srivastava, J. Biomol. Struct. Dyn.1-51, (2020).
30. D. Van Der Spoel, E. Lindahl, B. Hess, G. Groenhof, AE. Mark, HJ. Berendsen, J. Comput. Chem. 26, 1701-1718, (2005).
31. J. Prajapati, R. Patel, D. Goswami, M. Saraf, RM. Rawal, Comput. Biol. Med. 104568, (2021).
32. MJ. Abraham, T. Murtola, R. Schulz, S. Páll, JC. Smith, B. Hess, E. Lindahl, SoftwareX. 1, 19-25, (2015).
33. S. Keretsu, SP. Bhujbal, SJ. Cho, Sci. Rep. 10,1-14, (2020).
34. S. Verma, AK. Pandey, 3Biotech. 11, 1-10, (2021).
35. S. Pant, M. Singh, V. Ravichandiran, U. Murty, HK. Srivastava, J. Biomol. Struct. Dyn. 39, 2904-2913, (2020).
36. R. Ghosh, A. Chakraborty, A. Biswas, S. Chowdhuri, J. Mol. Struct. 1229, 129489, (2021).
37. MA. Said, A. Albohy, MA. Abdelrahman, HS. Ibrahim, Eur. J. Pharm. Sci. 160,105744, (2021).
38. P. Yadav, M. Rana, P. Chowdhury, J. Mol. Struct. 1246, 131253, (2021).
39. S. Keretsu, SP. Bhujbal, SJ. Cho, Sci. Rep. 9, 1-14, (2019).
40. D. Mishra, RR. Maurya, K. Kumar, NS. Munjal, V. Bahadur, S. Sharma, P. Singh, I. Bahadur, J. Mol. Liq. 335, 116185, (2021).
41. HJ. Berendsen, Jv. Postma, WF. van Gunsteren, A. DiNola, JR. Haak, J. Chem. Phys. 81, 3684-3690, (1984).
42. B. Hess, H. Bekker, HJ. Berendsen, JG. Fraaije, J. Comput. Chem. 18, 1463-1472, (1997).
43. M. Aldeghi, MJ. Bodkin, S. Knapp, PC. Biggin, J. Chem. Inf. Model. 57, 2203-2221,(2017).
44. S. Genheden, U .Ryde, Expert. Opin. Drug. Discov. 10, 449-461, (2015).
45. R. Kumari, R. Kumar, J. Chem. Inf. Model. 54, 10.1021, (2014).
46. TI. Adelusi, A-QK. Oyedele, OE. Monday, ID. Boyenle, MO. Idris, AT. Ogunlana, AM. Ayoola, JO. Fatoki, OE. Kolawole, KB. David, J. Mol. Struct.131879, (2021).
47. T. Topal, Y. Zorlu, N. Karapınar, J. Mol. Struct. 1239, 130514, (2021).
48. A. Daina, O. Michielin, V. Zoete, Sci. Rep. 7, 1-13, (2017).
49. MY. Al-Nour, MM. Ibrahim, T. Elsaman, Curr. Pharmacol. Rep. 5, 255-280, (2019).
50. M. Pooja, GJ. Reddy, K. Hema, S. Dodoala, B. Koganti, Eur. J. Pharmacol. 890,173688, (2021).
51. P. Chinnasamy, R. Arumugam, India. Egypt. J. Basic. Appl. Sci. 5, 265-279, (2018).
52. P. Sethi, Y. Bansal, G. Bansal, Med. Chem. Res. 27, 61-71, (2018).
53. S. Lau, P. Wang, B. Mok, A. Zhang, H. Chu, A. Lee, S. Deng, P. Chen, K. Chan, W. Song, Emerg. Microbes. Infect. 9, 837–842, (2020).