- COVID-19, SARS-CoV-2, Indian herbal plants, Natural compounds, Docking, ADMET analysis
Copyright (c) 2022 SChQ

This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License.
Abstract
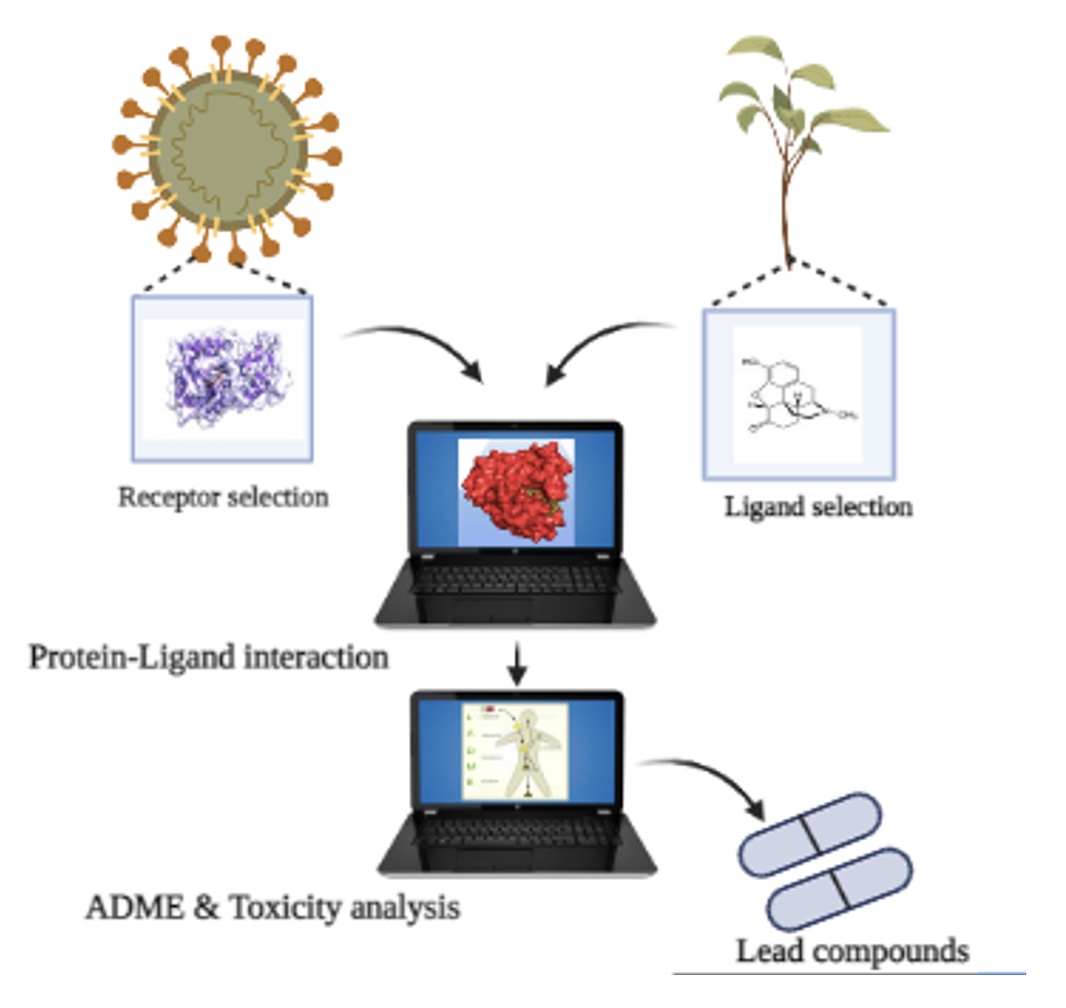
The novel coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) began in Wuhan, China, in December 2019 and quickly spread across the worldwide. It becomes a global pandemic and risk to the healthcare system of almost every nation around the world. In this study thirty natural compounds of 19 Indian herbal plants were used to analyze their binding with eight proteins associated with COVID -19. Based on the molecular docking as well as ADMET analysis, isovitexin, glycyrrhizin, sitosterol, and piperine were identified as potential herbal medicine candidates. On comparing the binding affinity with Ivermectin, we have found that the inhibition potentials of the Trigonella foenum-graecum (fenugreek), Glycyrrhiza glabra (licorice), Tinospora cordifolia (giloy) and Piper nigrum (black pepper) are very promising with no side-effects.
References
- World Health Organization (WHO). Novel Coronavirus (2019-nCoV) Situation Report – 52. Data as reported by 12 March 2020. Available from https://www.who.int/docs/default-source/coronaviruse/20200312-sitrep-52-covid-19.pdf?sfvrsn=e2bfc9c0_2 Accessed on 12 March 2020.
- N. Chen, M. Zhou, X. Dong, J. Qu, F. Gong, Y. Han, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 395, 507–13, (2020).
- J. Lan, J. Ge, J. Yu, S. Shan, H. Zhou, et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 581, 215–220, (2020).
- H. Hofmann, S. Pohlmann, Cellular entry of the SARS coronavirus. Trends Microbiol. 12, 466–472, (2004)
- H. Vennema, G.J. Godeke, J.W. Rossen, W.F. Voorhout, M.C. Horzinek, et al. Nucleocapsid-independent assembly of coronavirus-like particles by co-expression of viral envelope protein genes. EMBO J. 15, 2020–2028, (1996)
- Y.L. Siu, K.T. Teoh, J. Lo, C.M. Chan, F. Kien, et al. The M, E, and N structural proteins of the severe acute respiratory syndrome coronavirus are required for efficient assembly, trafficking, and release of virus-like particles. J Virol. 82, 11318–11330, (2008)
- Y. Gao, L. Yan, Y. Huang, F. Liu, Y. Zhao, et al. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 368, 779–782, (2020)
- R, Solecki, I.V. Shanidar, a Neanderthal flower burial in Northern Iraq. Science. 190, 880-1, (1975)
- B.A. Rasool Hassan, Medicinal plants (importance and uses). Pharmaceut Anal Acta. 3, 139, (2012)
- G. Samuelsson, Drugs of Natural Origin: A Textbook of Pharma¬cognosy, 5th Swedish Pharmaceutical Press, Stockholm. Swedish Phar¬maceutical Press, Stockholm. J Nat Prod, 68 (4), 631, (2004)
- J. Azmir, I.S.M. Zaidul, M.M. Rahman, K.M. Sharif, A. Mohamed, F. Sahena, M.H.A. Jahurul, K. Ghafoor, N.A.N. Norulaini, A.K.M. Omar. Techniques for extraction of bioactive compounds from plant materials: A review. Journal of Food Engineering. 117, 426-436, (2013)
- P.S. Masters, L.S. Sturman, Background Paper Functions of the Coronavirus Nucleocapsid Protein. In Coronaviruses and Their Diseases (Cavanagh, D. and Brown, T.D.K.eds), Springer US, Boston, MA, (1990)
- L. Subissi, C.C. Posthuma, J.C. Collet, Zevenhoven-Dobbe, A.E. Gorbalenya, E. Decroly, et al, One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc. Natl Acad. Sci U.S.A.111, E3900 (2014)
- Z.J. Miknis, E.F. Donaldson, T.C. Umland, R.A. Rimmer, R.S. Baric, L.W. Schultz, Severe acute respiratory syndrome coronavirus nsp9 dimerization is essential for efficient viral growth. J. Virol. 83, 3007–3018, (2009)
- R. Ulferts, J. Ziebuhr, Nidovirus ribonucleases: structures and functions in viral replication. RNA Biol. 8, 295–304, (2011)
- W. Rut, Z. Lv, M. Zmudzinski, S. Patchett, D. Nayak, S.J. Snipas, et al, Activity profiling and crystal structures of inhibitor-bound SARS-CoV-2 papain-like protease: A framework for anti -COVID-19 drug design. Science Advances 6, eabd4596, (2020)
- T. Muramatsu, Y.T. Kim, W. Nishii, T. Terada, M. Shirouzu, S. Yokoyama, Autoprocessing mechanism of severe acute respiratory syndrome coronavirus 3C-like protease (SARS-CoV 3CLpro) from its polyproteins FEBS J. 280, 2002–2013, (2013)
- P. Krafcikova, J. Silhan, R. Nencka, E. Boura, Structural analysis of the SARS-CoV-2 methyltransferase complex involved in RNA cap creation bound to sinefungin. Nat. Commun. 11, 3717, (2020)
- S.D. Conner, S.L. Schmid, Regulated portals of entry into the cell. Nature, 422, 37–44 (2003)
- S. Srivastava & A. Pandey, Computational screening of anticancer drugs targeting miRNA155 synthesis in breast cancer. Indian Journal of Biochemistry & Biophysics, 57, 389-394, (2020)
- I. Singha, S. Saxena, S. Gautam, A. Saha & S.K. Das, Grape extract protect against ionizing radiation-induced DNA damage. Indian J Biochem Biophys, 57, 219, (2020)
- A. Ganeshpurkar & A. Saluja, In silico interaction of rutin with some immunomodulatory targets: a docking analysis. Indian J Biochem Biophys, 55, 88, (2018)
- I.V. Tetko & V.Y. Tanchuk, Application of associative neural networks for prediction of lipophilicity in ALOGPS 2.1 program. J Chem Inf Comput Sci, 42, 1136, (2002)
- F. Cheng, W. Li, Y. Zhou, J. Shen, Z. Wu, G. Liu, P.W. Lee & Y. Tang, admetSAR: a comprehensive source and free tool for assessment of chemical ADMET properties. J Chem Inf Model, 52, 3099, (2012)
- A. Basu, A. Sarkar, U. Maulik & P. Basak, Three-dimensional structure prediction and ligand-protein interaction study of expansin protein ATEXPA23 from Arabidopsis thaliana L. Indian J Biochem Biophys, 56, 20, (2019)
- J. Blaising, S.J. Polyak, E.I. Pécheur, Arbidol as a broad-spectrum antiviral: An update. Antiviral Res. 107, 84–94, (2014)
- P. Gautret, J.C. Lagier, P. Parola, V.T. Hoang, L. Meddeb, et al, Hydroxychloroquine and azithromycin as a treatment of COVID‐19: results of an open‐label non‐randomized clinical trial. Int J Antimicrob Agents – In Press 17 March 2020.
- L. Caly, J.D. Druce, M.G. Catton, D.A. Jans, K.M. Wagstaff, The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in Vitro. Antiviral Research, 178, 104787, (2020)
- ClinicalTrials.gov [Internet]. Bethesda (MD): National Library of Medicine (US). Mar 12 – Identifier NCT04252885, The efficacy of lopinavir plus ritonavir and arbidol against novel coronavirus infection (ELACOI), (2020)
- ClinicalTrials.gov [Internet]. Bethesda (MD): National Library of Medicine (US). Mar 12 – Identifier NCT04257656, Severe2019-nCoV Remdesivir RCT, (2020)
- S.K. Bhal, LogP—making sense of the value. Advanced Chemistry Development, (Toronto, ON, Canada), 1, (2007)
- W.L. Jorgensen, & E.M. Duffy, Prediction of drug solubility from Monte Carlo simulations. Bioor. Med. Chem. Lett. 10, 1155-1158, (2000)
- F. Cheng, W. Li, Y. Zhou, J. Shen, Z. Wu, G. Liu, P.W. Lee & Y. Tang, admetSAR: a comprehensive source and free tool for assessment of chemical ADMET properties. J Chem Inf Model, 52, 3099, (2012)