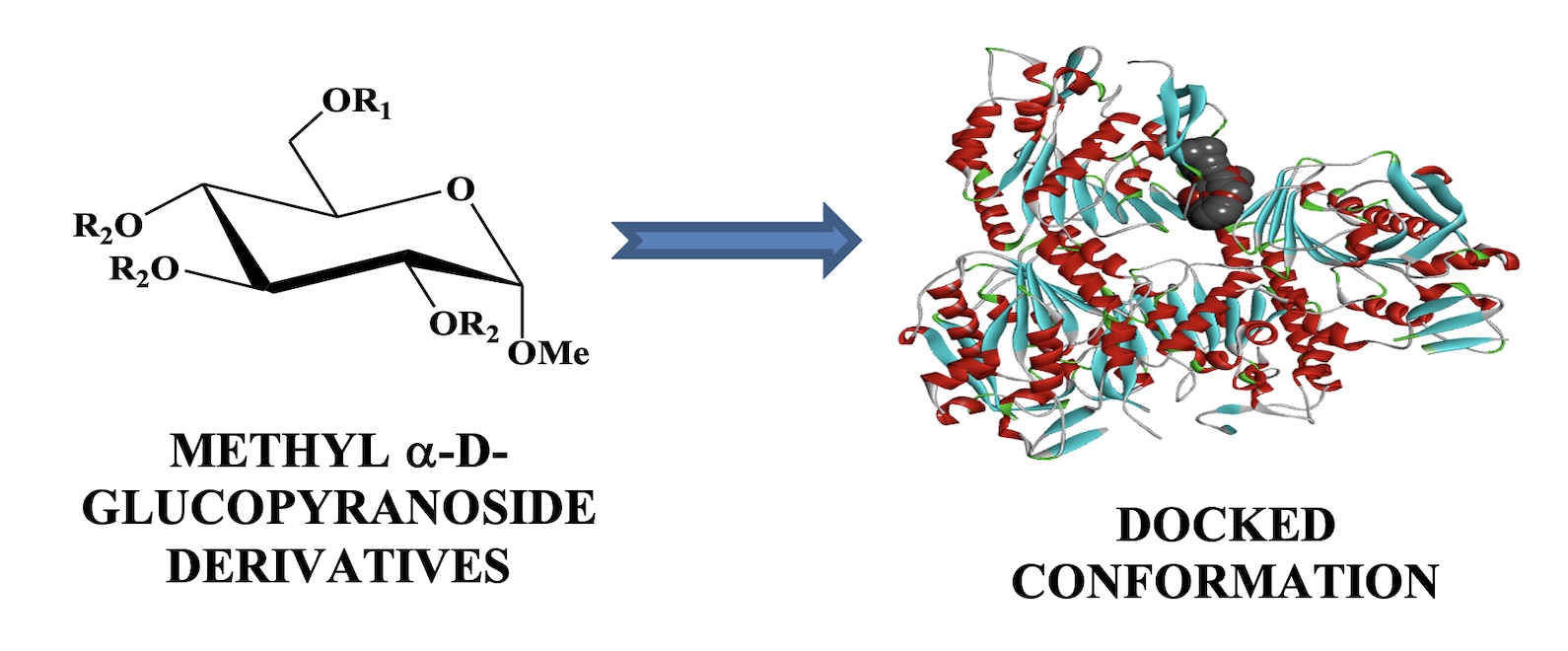
COMPUTATIONAL INVESTIGATION OF METHYL α-D-GLUCOPYRANOSIDE DERIVATIVES AS INHIBITOR AGAINST BACTERIA, FUNGI AND COVID-19 (SARS-2)

- Glucopyranoside, ADMET, Molecular Docking, Drug-likeness, Toxicity, Amino Acids
Copyright (c) 2021 SChQ

This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License.
Abstract
For employing computational tools for drug discovery in the area of medicinal chemistry by carbohydrates, methyl a-D-glucopyranoside and its ten acylated derivatives have picked up. At first, the HOMO, LUMO, and its energy gap have been obtained by the DFT method, as well as the chemical reactivity and global descriptors, such as global softness, electron affinity, ionization potential, electronegativity, global hardness, global electrophilicity index, and chemical potential have calculated from HOMO and LUMO data. From this data, it is illustrated that the HOMO-LUMO energy gap is -9.756 to -7.756 kcal/mol while the compound 12 shows the highest energy gap and compound 10 is opposite, and the softness has recorded the range from 0.208 to 0.255, showing a small difference, while the lower softness is picked up for 12, but 09 is reverse. The key and vital part of this study are noted as molecular docking against four pathogens proteins, such as Bacillus cereus, E Coli, Lanosterol 14alpha demethylase, SARS-02, and it is obtained the most expected and impactful result as an inhibitor. It is mentioned that the result of molecular docking score is -9.6 for compounds 09 and 11 against Bacillus cereus which is the highest score. But it is slightly different for E Coli -9.5 and -9.3 of compounds 10 and 07. On the other hand, it is precious and lavish work against COVID-19 protein whereas all of the tested compounds can show good and standard inhibitor with value more than -6.0, and the -9.1, 9.0, and 8.5 for compounds 09, 07, and 08, respectively. It may be revealed that methyl α-D-glucopyranoside and its ten acylated derivatives are also found as a good inhibitor against SARS-02 protein than bacteria and fungi. Moreover, all of these are non–carcinogenic and low toxic in the case of both aquatic and non-aquatic species which says us for safe use in drug discovery.
References
- P. K. Hohenberg, W, Phys. Rev.136, B864, (1964).
- W. B. Kohn, D. Axel, Parr, G. Robert, J.Phys. Chem. 100, 12974-12980, (1996).
- G. K. Sliwoski, K. Sandeepkumar, J. Meiler, W. E. Lowe, Pharmacol. Rev. 66, 334-395, (2014).
- M. Zheng, L. Xian, X. Yuan, L. Honglin, L. Cheng, J. Hualiang, Trends pharmacol. sci. 34, 549-559, ( 2013).
- Z. Afroza, K. Ajoy, S. M. Nuruzzaman, P. Sunanda, Int. J. Chem. Technol. 3, 151-161, (2019).
- K. Ajoy, P. Sunanda, S. M. Nuruzzaman, M. I. Jahidul, Int. J. Chem. Technol. 6, 236-253, (2019).
- M. I. Jahidul, S. M. Nuruzzaman, K. Ajoy; P. Sunanda, Int. J. Adv. Biol. Biomed. Res. 7, 318-337, (2019).
- M. I. Jahidul, K. Ajoy, S. M. Nuruzzaman, P. Sunanda, Z. Afroza, Adv. J. Chemi. 2, 316-326, (2019).
- M. J. Islam, S. M. Nuruzzaman, K. Ajoy, P. Sunanda, Int. J. Adv. Biol. Biomed. Res. 7, 318-337, (2019).
- K. Ajoy, S. M. Nuruzzaman, P. Sunanda, Turkish Comp. Theo. Chem. 3(2), 59-68, (2019).
- K. Ajoy, S. M. Nuruzzaman, P. Sunanda, Eurasian J. Environ. Res. 3, 1-10, (2019).
- R. L. Andrew, Pearson Education Limited, 2001.
- J. G. Vinter, G. Mark, Mol. Model. drug design. Macmillan International Higher Education, (1994).
- A. Daniel G. schwend, C. Andrew, I. D. Kuntz, J. Mol. Recognit. 9, 175-186, (1996).
- W. P. Walters, M. A. Ajay , M. A. Murcko, Curr. Opin. Chem. Biol. 3, 384-387, (1999).
- H. C. Kolb, K. S. Barry, Drug Discov. Today. 8, 1128-1137, (2003).
- L. H. Naylor, Biochem. Pharmacol. 58, 749-757, (1999).
- H. V. D. Waterbeemd, G. Eric, Nat. Rev. Drug Discov. 2, 192-204, (2003).
- J. A. Arnott, S. L. Planey, Exp. Opin. Drug Discov. 7, 863-875, (2012).
- K. Vimala, K. S. Samba, Y. M. Mohan, B. Sreedhar, K. M. Raju, Carbohydr. Polym. 75, 463-471, (2009).
- C. Rafin, V. Etienne, M. Sancholle, D. Postel, C. Len, P. Villa, G. Ronco, J. Agric. Food Chem. 48, 5283-5287, (2000).
- F. Cedeno-Laurent, J. O. Matthew, S. R. Barthel, D. Hays, T. Schatton, Q. Zhan, H. Xiaoying, K. L. Matta, J. G. Supko, M. H. Frank, J. Invest. Dermatol. 132, 410-420, (2012).
- J. E. Barradas, M. I. Errea, N.B. D’Accorso, C. S. Sepúlveda, L. B. Talarico, E. B. Damonte, Carbohydr. Res. 343, 2468-2474, (2008).
- L. Han, S. Wenlong, X. Li, A. Sik, H. Lin, K. Liu, L. Wang, Med. Chem. Comm. 10, 598-605, (2019).
- L. Gómez-García, R.-M. Irma, M. Rodríguez-Sosa, L. I. Terrazas, Parasitol. Res. 99, 440-448, (2006).
- E. Kerasioti, S. Dimitrios, A. Jamurtas, A. Kiskini, Y. Koutedakis, N. Goutzourelas, S. Pournaras, A. M. Tsatsakis, D. Kouretas, Food chem toxicol. 61, 42-46, (2013).
- T. K. Pal, D.Tuli, A. Chakrabarty, D. Dey, S. K. Ghosh, T. Pathak, Bioorg. Med. Chem. Lett. 20, 3777-3780, (2010).
- M. Islam, M. Arifuzzaman, M. Rahman, M. A. Rahman, S. M. A. Kawsar, Hacett. J. Biol. Chem. 47, 153-164, (2019).
- S. M. A. Kawsar, M. Islam, S. Jesmin, M. A. Manchur, I. Hasan, S. Rajia, Int. J. Biosci. 12, 408-416, (2018).
- S. M. A Kawsar, A. K. M. S. Kabir, M. M. Manik, M. K. Hossain, M. N. Anwar, Int. J. Biosci, 2, 66-73, (2012).
- A. Kabir, P. Dutta, M. N. Anwar, Pak. J. Biol. Sci. 7, 1730-1734, (2004).
- A. D. Becke, J. Chem. phys. 96, 2155-2160, (1992).
- R. G. Parr, Horizons of Quantum Chemistry, ed: Springer, 1980, pp. 5-15.
- R. G. Parr, Y. Weitao, J. Am. Chem. Soc. 106, 4049-4050, (1984).
- M. J. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, A. Robb, J. R. Cheeseman, G. Scalmani, V. Barone, B. Mennucci, G. A. Petersson , Gaussian 09. Gaussian Inc, Wallingford CT, 93, 2009.
- W. J. Hehre, S. F. Robert, J. A. Pople, J. Chem. Phys. 51, 2657-2664, (1969).
- C. Sosa, A. Jan, C. Lee, J. F. Blake, B. L. Chenard, T. W. Butler, Int. J. Quantum Chem. 49, 511-526, (1994).
- W. L. DeLano, "The PyMOL user's manual," http://www.pymol.org, 2002.
- O. Trott, O. J. Arthur, J. Comput. Chem. 31, 455-461, (2010).
- A. S. Inc, "Discovery Studio Modeling Environment, release 4.0," 2013.
- F. Cheng, L. Weihua, Y. Zhou, J. Shen, Z. Wu, G. Liu, P. W. Lee, Y. Tang, admetSAR: a comprehensive source and free tool for assessment of chemical ADMET properties, ed: ACS Publications, 2012.
- A. Daina, M. Olivier, V. Zoete, "SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules," Sci. Rep. 7, 42717, (2017).
- M. J. Islam, K. Ajoy, P. Sunanda, S. M. Nuruzaman, Chem. Methodol. 4, 130-142, (2020).
- A. Kumer, S. M. Nuruzaman, S. Paul, Turkish Comp. Theo. Chem. 3, 59-68, (2019).
- R. G. Parr, C. K. Pratim, J. Am. Chem. Soc. 113, 1854-1855, (1991).
- R. G. Parr, L. V. Szentpály, L. Shubin, J. Am. Chem. Soc. 21, 1922–24, (1999).
- C. A. Lipinski, L. Franco, B. W. Dominy, J. P. Feeney, Adv. Drug Deliv. Rev. 23, 3-25, (1997).
- N. J. Kumar, S. S. Mishara, H. P. Singh, S. Ranjan, C. S. Sharma. Int. J. Pharm. Sci. Drug Res. 10, 278-282, (2018).
- M. M. Hoque, K. Ajoy, H. M. Sajib, K. M. Wahab, Int. J. Adv. Biol. Biomed. Res. 9, 77-104, (2021).
- K. Fent, W. A. Anna, D. Caminada, Aquat. Toxicol. 76, 122-159, (2006).