POTENTIAL OF CURATELLA AMARICANA L. AGAINST SARS-COV2: BIOAVAILABILITY, MOLECULAR SIMILARITY AND MOLECULAR DOCKING BETWEEN SECONDARY METABOLITES AND PROTEASE TYPE 3-CHYMOTRYPSIN (3CLPRO)

- SARS-Cov2,
- secondary metabolites,
- Curatella amaricana L
Copyright (c) 2021 SChQ

This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License.
Abstract
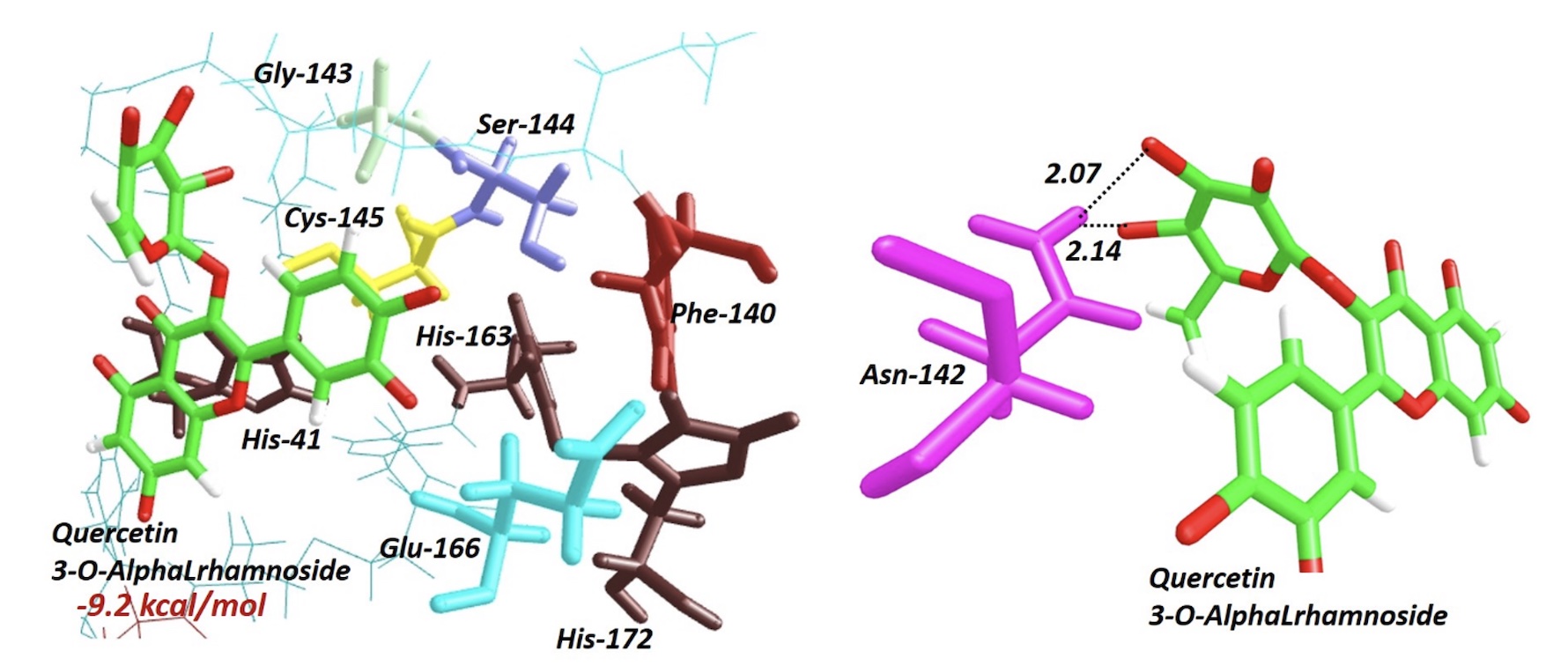
We report the bioavailability analysis for six secondary metabolites with antiviral, antioxidant and antitumor activity reported, from Curatella amaricana L. Additionally, the molecular similarity analysis of each metabolite is presented and compared with Lopinavir, Ritonavir, Darunavir, Cobicistat and Nelfinavir, which actually are in the third phase for the production of a new vaccine for SARS-Cov2. The mode of interaction through molecular docking between each structure and the zone of action for protease type 3-chymotrypsin (3CLpro) also is presented. The molecular geometry for structures were optimized at semiempirical PM6 level. The bioavailability and molecular docking calculations were performed using the algorithms incorporated in chemoinformatic servers and AutoDock Vina. The results show that the structures studied lead a moderated permeability through the cell membrane, by complying with Lipinski's "rule of 5”. Molecular similarity was evaluated by averaging geometric parameters (3D-Shape) and electrostatic potential (ESP). The results show that the most secondary metabolites would have a similar mode of action as the Lopinavir, with average similarity between 0.65 and 0.73. This last idea is reinforced by the results for molecular docking with the 3CLpro active site, highlighting the interaction of the molecules studied with the amino acid residues: His-41, Phe-140, Gly-143, Ser-144, Cys-145, His-163, Glu-166 and His-172, with an range interaction-free energy between -7.2 kcal/mol and -9.2 Kcal/mol, highlighting Quercetin 3-O-Alpha-L-rhamnoside with improve affinity energy than Lopinavir.
References
- G. L. Clercq and E. De, "Therapeutic options for the 2019 novel coronavirus (2019-nCoV)," Nat. Rev. Drug Discov, vol. 19, pp. 149-150, 2020. DOI: https://10.1038/d41573-020-00016-0
- Yu Wai Chen, Chin-Pang Bennu Yiu and Kwok-Yin Wong, "Prediction of the SARS-CoV-2 (2019-nCoV) 3C-like protease (3CLpro) structure: virtual screening reveals velpatasvir, ledipasvir, and other drug repurposing candidates," F1000 Research, vol. 9, p. 129, 2020. DOI: https://10.12688/f1000research.22457.2
- P. G. Taylor, I. M. Cesari, M. Arsenak, D. Ballen, M. J. Abad, A. Fernández, B. Milano, M.-C. Ruiz and B. W. &. F. Michelangeli, "Evaluation of Venezuelan Medicinal Plant Extracts for Antitumor and Antiprotease Activities," Pharm. Biol., vol. 44, pp. 349-362, 2006. DOI: https://doi.org/10.1080/13880200600748119
- R. Duno de Stefano, G. Aymard and O. Huber, Flora Vascular de los Llanos de Venezuela, Caracas . Fundación Empresas Polar - FUNDENA – FIBV, 2007, p. 738 páginas.
- A. Bender and R. C. Glen, "Molecular similarity: a key technique in molecular informatics," Org. Biomol. Chem., vol. 2, pp. 3204-3218, 2004. DOI: https://doi.org/10.1039/B409813G
- H. Eckert and J. Bajorath, "Molecular similarity analysis in virtual screening: foundations, limitations and novel approaches," Drug Discov Today, vol. 12, pp. 225-233, 2007. DOI: https://doi.org/10.1016/j.drudis.2007.01.011
- S. Jo, S. Kim and D. H. S. &. M.-S. Kim, "Inhibition of SARS-CoV 3CL protease by flavonoids," J. Enzyme Inhib., vol. 35, pp. 145-151, 2020. DOI: https://doi.org/10.1080/14756366.2019.1690480
- M. W. Schmidt, K. K. Baldridge, J. A. Boatz, S. T. Elbert, M. S. Gordon, J. H. Jensen, S. Koseki, N. Matsunaga, K. A. Nguyen, S. Su, T. L. Windus, M. Dupuis and J. A. Montgomery J, "General Atomic and Molecular Electronic Structure System," Comput. Chem., vol. 14, pp. 1347-1363, 1993. DOI: https://doi.org/10.1002/jcc.540141112
- M. J. Vainio, J. S. Puranen and M. S. Johnson, "ShaEP: molecular overlay based on shape and electrostatic potential.," J Chem Inf Model., vol. 49, pp. 492-502., 2009. DOI: https://doi.org/10.1021/ci800315d
- W.-H. Shin, Z. Xiaolei, M. G. Bures and D. Kihara, "Three-Dimensional Compound Comparison Methods and Their Application in Drug Discovery," Molecules, vol. 20, pp. 12841-12862, 2015. DOI: https://doi.org/10.3390/molecules200712841
- I. Muegge, "Selection Criteria for Drug-Like Compounds," Med.l Res. Rev, vol. 23, pp. 302-321, 2003. DOI: https://doi.org/10.1002/med.10041
- C. A. Lipinski, F. Lombardo, B. W. Dominy and P. J. Feeney, "Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings," Adv. Drug Deliv. Rev, vol. 15, pp. 3-25, 1997. DOI: https://doi.org/10.1016/S0169-409X(96)00423-1
- Molinspiration , "Cheminformatics on the Web," 1986. [Online]. Available: http://www.molinspiration.com/.
- T.-W. Lee, M. M. Cherney, J. Liu, K. E. James, J. C. Powers, L. D. Eltis and M. N. James, "Crystal Structures Reveal an Induced-fit Binding of a Substrate-like Aza-peptide Epoxide to SARS Coronavirus Main Peptidase," J. Mol. Biol., vol. 366, pp. 916-932, 2007. DOI: https://doi.org/10.1016/j.jmb.2006.11.078
- A. Pedretti, L. Villa and G. Vistoli, "VEGAZZ: an open platform to develop chemo-bio-informatics applications, using plug-in architecture and script programming," J Comput Aided Mol Des, vol. 18, pp. 167-173 , 2004.DOI: https://doi.org/10.1023/B:JCAM.0000035186.90683.f2
- J. Gasteiger and M. Marsili, "Iterative partial equalization of orbital electronegativity—a rapid access to atomic charges," Tetrahedron, vol. 36, pp. 3219-3228, 1980. DOI: https://doi.org/10.1016/0040-4020(80)80168-2
- H. Zakaryan, E. Arabyan, A. Oo and K. Zandi, "Flavonoids: promising natural compounds against viral infections.," Arch. Virol., vol. 162, p. 2539–2551, 2017. DOI: https://doi.org/10.1007/s00705-017-3417-y
- Y. B. Ryu, H. J. Jeong, J. H. Kim, Y. M. Kim, J.-Y. Park, D. Kim, T. T. H. Naguyen, S.-J. Park, J. S. Chang, K. H. Park, M.-C. Rho and W. S. Lee, "Biflavonoids from Torreya nucifera displaying SARS-CoV 3CLpro inhibition," Bioorg. Med. Chem., vol. 18, pp. 7940-7947, 2010. DOI: https://doi.org/10.1016/j.bmc.2010.09.035
- H. Pajouhesh and G. R. Lenz, "Medicinal Chemical Properties of Successful Central Nervous System Drugs," NeuroRx, vol. 2, p. 541–553, 2005. DOI: https://doi.org/10.1602/neurorx.2.4.541
- S. A. Hitchcock and L. D. Pennington, "Structure−Brain Exposure Relationships," J. Med. Chem., vol. 49, pp. 7559-7583, 2006. DOI: https://doi.org/10.1021/jm060642i
- D. Veber, S. Johnson, H. Cheng, B. Smith, K. Ward and K. Kopple, " Molecular properties that influence the oral bioavailability of drug candidates," J Med Chem, vol. 45, pp. 2615-2623, 2002. DOI: https://doi.org/10.1021/jm020017n
- A. Daina and Z. Vincent, "A BOILED-Egg To Predict Gastrointestinal Absorption and Brain Penetration of Small Molecules," ChemMedChem, vol. 11, p. 1117–1121, 2016. DOI: https://doi.org/10.1002/cmdc.201600182
- A. Daina, O. Michielin and V. Zoete, "SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules.," Sci. Rep., vol. 7, p. 42717, 2017. DOI: https://doi.org/10.1038/srep42717
- D. M, Taleb-Gassabi and D. M, "Lopinavir; A Potent Drug against Coronavirus Infection: Insight from Molecular Docking Study," Arch. Clin. Infect. Dis., vol. 12, p. e13823, 2017. DOI: 10.5812/archcid.13823
- L. Deng, C. Li, Q. Zeng, X. Liu, X. Li, H. Zhang, Z. Hong and J. Xia, "Arbidol combined with LPV/r versus LPV/r alone against Corona Virus Disease 2019: A retrospective cohort study," J. Infect., vol. Article in press, 2020. DOI: https://doi.org/10.1016/j.jinf.2020.03.002
- J. Cui, X. Liu and L. M. Chow., "Flavonoids as P-gp Inhibitors: A Systematic Review of SARs," Curr. Med. Chem. vol. 26, pp. 4799-4831, 2019. DOI: 10.2174/0929867325666181001115225
- N. Yamamoto, R. Yang, Y. Yoshinaka, S. Amari, T. Nakano, J. Cinatl, H. Rabenau, H. W. Doerr, G. Hunsmann, A. Otaka, H. Tamamura, N. Fujii and N. Yamamoto, "HIV protease inhibitor nelfinavir inhibits replication of SARS-associated coronavirus," Biochem. Biophys. Res. Commun., vol. 318, pp. 719-725, 2004. DOI: https://doi.org/10.1016/j.bbrc.2004.04.083
- T. Sheahan, A. Sims and S. Leist, "Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV," Nat. Commun., vol. 11, p. 222, 2020. DOI: https://doi.org/10.1038/s41467-019-13940-6
- S. K. Bhal, "LogP-Making Sense of the Value," 2012. [Online]. Available: http://www.acdlabs.com/logp.