CONVERSION OF LINOLEIC ACID TO DIFFERENT FATTY ACID METABOLITES BY LACTOBACILLUS PLANTARUM 13-3 AND IN SILICO CHARACTERIZATION OF THE PROMINENT REACTIONS
- Lactobacillus plantarum,
- fatty acid metabolites,
- Linoleate isomerase,
- Acetoacetate decarboxylase,
- Dehydrogenase
- in silico study ...More
Copyright (c) 2020 SChQ

This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License.
Abstract
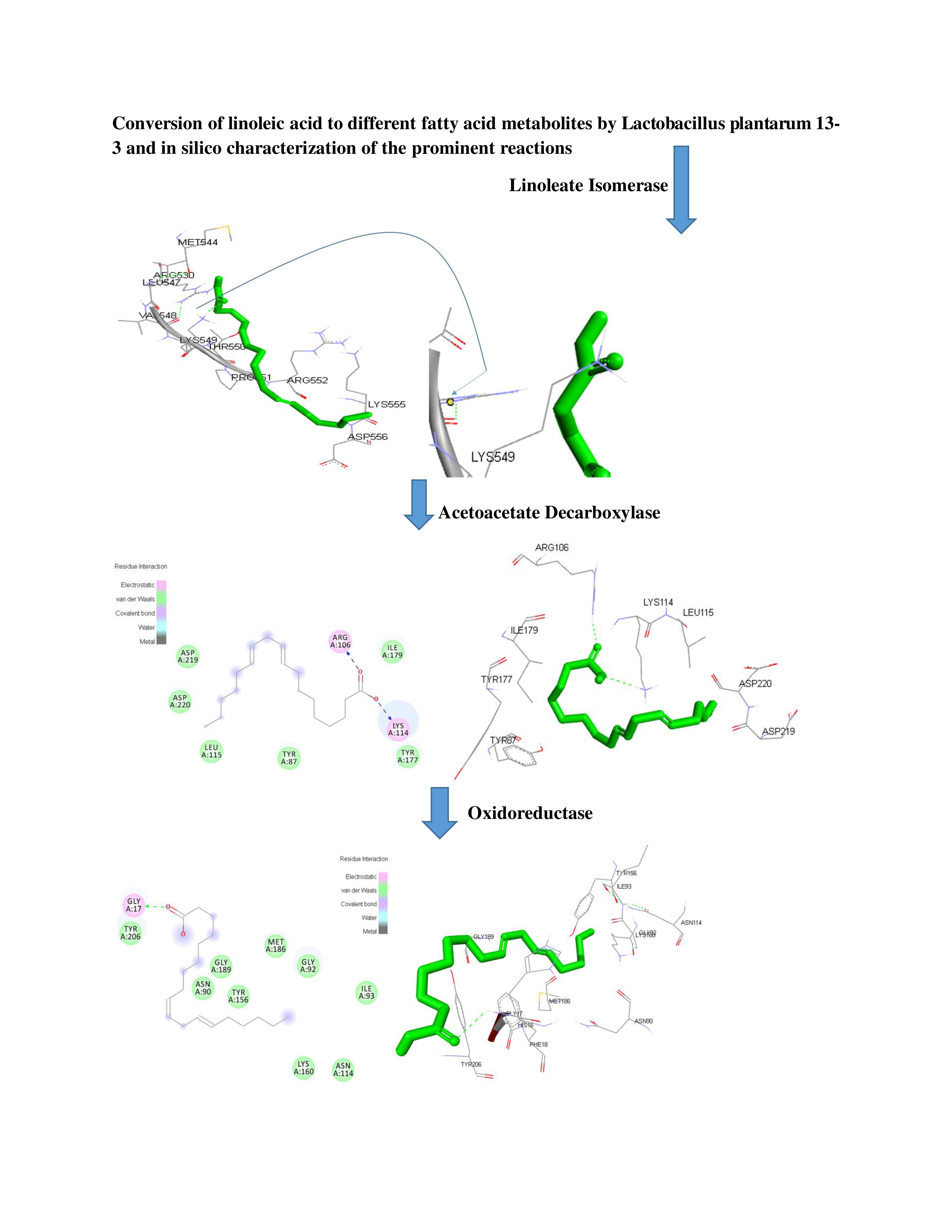 Lactobacillus plantarum strains have been extensively used in food processing and preservation. L. plantarum also has the potential to convert polyunsaturated fatty acids, e.g. linoleic acid (LA) into bioactive and less toxic fatty acid metabolites. The objective of this study was to assess the capability of probiotic L. plantarum 13-3 to convert Linoleic Acid (LA) to different fatty acid metabolites in the medium supplemented with differential concentrations of LA, and the relevant reactions were characterized by in silico calculation. L. plantarum 13-3 was grown in MRS medium containing LA from 1% to 10%, and the fatty acid metabolites formed in the medium were identified and quantitated by GC-MS and in silico studies were done to confirm the enzymatic reactions involved in its conversion. The results showed that L. plantarum 13-3 could convert LA at different concentrations to 5 different fatty acid metabolites i.e, (Z)-Ethyl heptadec-9-enoate, 9,12-Octadecadienoic acid (Z, Z), methyl ester, Octadec-9-enoic acid, cis-11,14-Eicosadienoic acid, methyl ester and (Z)-18-Octadec-9-enolide. Among these metabolites, the formation of an long chain fatty acid Octadec-9-enoic Acid, also known as 18:1, N-9 or Delta(9)-Octadecenoic acid, is classified as a member of the Long-chain fatty acids in media supplemented with 4% to 10% LA, is being reported for the first time. Putative candidate enzymes involved in biotransformation of LA into fatty acid metabolites were identified in whole genome of L. plantarum 13-3, sequenced previously. In silico studies confirmed that several enzymes including the linoleate isomerase, acetoacetate decarboxylase and oxidoreductase may be involved in biotransformation of LA into fatty acid metabolites. These enzymes could effectively bind the LA molecule mainly by forming hydrogen bonding between the acidic groups of LA and the proline residues at the active sites of the enzymes validating the putative reacting partners.
Lactobacillus plantarum strains have been extensively used in food processing and preservation. L. plantarum also has the potential to convert polyunsaturated fatty acids, e.g. linoleic acid (LA) into bioactive and less toxic fatty acid metabolites. The objective of this study was to assess the capability of probiotic L. plantarum 13-3 to convert Linoleic Acid (LA) to different fatty acid metabolites in the medium supplemented with differential concentrations of LA, and the relevant reactions were characterized by in silico calculation. L. plantarum 13-3 was grown in MRS medium containing LA from 1% to 10%, and the fatty acid metabolites formed in the medium were identified and quantitated by GC-MS and in silico studies were done to confirm the enzymatic reactions involved in its conversion. The results showed that L. plantarum 13-3 could convert LA at different concentrations to 5 different fatty acid metabolites i.e, (Z)-Ethyl heptadec-9-enoate, 9,12-Octadecadienoic acid (Z, Z), methyl ester, Octadec-9-enoic acid, cis-11,14-Eicosadienoic acid, methyl ester and (Z)-18-Octadec-9-enolide. Among these metabolites, the formation of an long chain fatty acid Octadec-9-enoic Acid, also known as 18:1, N-9 or Delta(9)-Octadecenoic acid, is classified as a member of the Long-chain fatty acids in media supplemented with 4% to 10% LA, is being reported for the first time. Putative candidate enzymes involved in biotransformation of LA into fatty acid metabolites were identified in whole genome of L. plantarum 13-3, sequenced previously. In silico studies confirmed that several enzymes including the linoleate isomerase, acetoacetate decarboxylase and oxidoreductase may be involved in biotransformation of LA into fatty acid metabolites. These enzymes could effectively bind the LA molecule mainly by forming hydrogen bonding between the acidic groups of LA and the proline residues at the active sites of the enzymes validating the putative reacting partners.
References
- Wang R, Kern JT, Goodfriend TL, Ball DL, Luesch H. Activation of the antioxidant response element by specific oxidized metabolites of linoleic acid. Prostaglandins Leukot. Essent. Fatty Acids. 81, 5359, (2009).
- Cunnane SC, Guesnet P. Linoleic acid recommendations-a house of cards. Prostaglandins Leukot. Essent. Fatty Acids. 85, 399402, (2011).
- Mathieu P, Pibarot P, Despres JP. Metabolic syndrome: the danger signal in atherosclerosis. Vasc. Health Risk Manag .2, 285302, (2006).
- Niki E. Lipid peroxidation: physiological levels and dual biological effects. Free Radic. Biol. Med. 47, 469484, (2009).
- Tavakoli Yaraki, M Karami Tehrani F. Apoptosis Induced by 13-hydroxyoctadecadienoic acid in the breast cancer cell lines, MCF-7 and MDA-MB-231. Iran. J. Basic Med. Sci. 16, 653659, (2013).
- Vangaveti V, Baune BT, Kennedy RL. Hydroxyoctadecadienoic acids: novel regulators of macrophage differentiation and atherogenesis. Ther. Adv. Endocrinol. Metab. 1, 5160, (2010).
- Yuan H, Li MY, Ma LT, et al. 15-Lipoxygenases and its metabolites 15(S)-HETE and 13(S)-HODE in the development of non-small cell lung cancer Thorax 65, 321326, (2010).
- J.F. Greene, B.D. Hammock, Toxicity of linoleic acid metabolites, in: K.V. Honn, L.J. Marnett, S. Nigam, C.N. Serham, E.A. Dennis (Eds.), Eicosanoids and Other Bioactive Lipids in Cancer, Inflammation, and Radiation Injury, Kluwer Academic/Plenum Press, New York. 471477, (1999).
- M.R. Buchanan, Linoleic acid metabolites in health and disease, Adv. Exp. Med. Biol. 469, 463469, (1999).
- Sonnenburg, JL.; Backhed, F. Diet-microbiota interactions as moderators of human metabolism. Nature, 535, 7610,664, (2016).
- Marc Schoeler.; Robert Caesar. Dietary lipids, gut microbiota and lipid metabolism Reviews in Endocrine and Metabolic Disorders 112, (2019).
- DL, Greenway.; KG, Dyke. Mechanism of the inhibitory action of linoleic acid on the growth of Staphylococcus aureus. J. Gen. Microbiol. 115, 233245, (1979).
- MK, Raychowdhury.; R, Goswami.; P, Chakrabarti. Effect of unsaturated fatty acids in growth inhibition of some penicillin-resistant and sensitive bacteria, J. Appl. Bacteriol. 59, 183188, (1985).
- H. Keweloh.; H.J. Heipieper. Trans unsaturated fatty acids in bacteria, Lipids 31, 129137, (1966).
- CJ, Zheng.; JS Yoo.; TG Lee.; HY Cho.; YH Kim.; WG Kim. Fatty acid synthesis is a target for antibacterial activity of unsaturated fatty acids. FEBS Lett. 579, 51575162, (2005).
- A, Buccioni.; M, Decandia.; S, Minieri.; G, Molle.; A, Cabiddu. Lipid metabolism in the rumen: new insights on lipolysis and biohydrogenation with an emphasis on the role of endogenous plant factors, Anim. Feed Sci. Technol. 174,125, (2012).
- MR, Maia LC.; Chaudhary.; L, Figueres.; RJ, Wallace. Metabolism of polyunsaturated fatty acids and their toxicity to the microflora of the rumen. Antonie Van Leeuwenhoek 91, 303314, (2007).
- N, McKain.; KJ, Shingfield.; RJ, Wallace. Metabolism of conjugated linoleic acids and 18: 1 fatty acids by ruminal bacteria: products and mechanisms, Microbiol-Sgm. 156579–588, (2010).
- IS, Nam.; PC, Garnsworthy. Biohydrogenation of linoleic acid by rumen fungi compared with rumen bacteria. J. Appl. Microbiol. 103, 551556, (2007).
- Selhub EM, Logan AC, Bested AC, Fermented foods, microbiota, and mental health: ancient practice meets nutritional psychiatry. J. Physiol. Anthropol. 33, 112, (2014).
- Shah NP, Functional cultures and health benefits. Int. Dairy J.17, 12621277, (2007).
- Younesi E, Ayseli MT. An integrated systems-based model for substantiation of health claims in functional food development. Trends Food Sci. Tech. 41,95100, (2015)
- S. Torriani, F. Clementi, M. Vancanneyt, B. Hoste, F. Dellaglio, and K. Kersters, “Differentiation of Lactobacillus plantarum, L. pentosus and L. paraplantarum species by RAPD-PCR and AFLP,” Systematic and Applied Microbiology, 24, 4, 554560, (2001).
- E. A. Pfeiler and T. R. Klaenhammer, “The genomics of lactic acid bacteria,” Trends in Microbiology, 15, 12, 546553, (2007).
- R. J. Siezen, V. A. Tzeneva, A. Castioni et al., “Phenotypic and genomic diversity of Lactobacillus plantarum strains isolated from various environmental niches,” Environmental Microbiology, 12,3, 758773, (2010).
- R. J. Siezen and J. E. T. van Hylckama Vlieg, “Genomic diversity and versatility of Lactobacillus plantarum, a natural metabolic engineer,” Microbial Cell Factories. 10, S3, 1, (2011).
- J. Wang, H. Ji, D. Zhang et al., “Assessment of probiotic properties of Lactobacillus plantarum ZLP001 isolated from gastrointestinal tract of weaning pigs,” African Journal of Biotechnology. 10,54, 1130311308, (2011).
- Y. Nami, N. Abdullah, B. Haghshenas, D. Radiah, R. Rosli, and A. Y. Khosroushahi, “Assessment of probiotic potential and anticancer activity of newly isolated vaginal bacterium Lactobacillus plantarum 5BL,” Microbiology and Immunology, 58, 9, 492502, (2014).
- R. Tabasco, F. Sánchez-Patán, M. Monagas et al., “Effect of grape polyphenols on lactic acid bacteria and Bifidobacteria growth: resistance and metabolism,” Food Microbiology, 28,7,13451352, (2011).
- P. Fras, F. M. Campos, T. Hogg, and J. A. Couto, “Production of volatile phenols by Lactobacillus plantarum in wine conditions. Biotechnology Letters. 36, 2, 281285, (2014).
- E. J. Yang and H. C. Chang, “Purification of a new antifungal compound produced by Lactobacillus plantarum AF1 isolated from kimchi,” International Journal of Food Microbiology. 139,12,5663, (2010).
- P. Li, Q. Zhou, and Q. Gu, “Complete genome sequence of Lactobacillus plantarum LZ227, a potential probiotic strain producing B-group vitamins,” Journal of Biotechnology. 234, 6670, (2016).
- K. B. Ahn, J. B. Baik, O. Park, C. Yun, and S. H. Han, “Lactobacillus plantarum lipoteichoic acid inhibits biofilm formation of Streptococcus mutans,” PLoS One. 13, 2, e0192694, (2018).
- S, Kishino.; J, Ogawa.; K, Yokozeki.; S, Shimizu. Linoleic acid isomerase in Lactobacillus plantarum AKU1009a proved to be a multi-component enzyme system requiring oxidoreduction cofactors. Biosci. Biotechnol. Biochem. 75,318322, (2011a).
- S. Kishino.; SB, Park.; M, Takeuchi.; K, Yokozeki.; S, Shimizu.; J, Ogawa. Novel multicomponent enzyme machinery in lactic acid bacteria catalyzing C=C double bond migration useful for conjugated fatty acid synthesis. Biochem. Biophys. Res. Commun. 416: 188193, (2011b).
- S, Kishino.; M, Takeuchi.; SB, Park.; A, Hirata.; N, Kitamura.; J, Kunisawa.; H, Kiyono.; R, Iwamoto.; Y, Isobe.; M, Arita.; H, Arai.; K, Ueda.; J, Shima.; S, Takahashi.; K, Yokozeki.; S, Shimizu.; J, Ogawa. Polyunsaturated fatty acid saturation by gut lactic acid bacteria affecting host lipid composition. Proc. Natl. Acad. Sci. USA. 110, 1780817813, (2013).
- Barrett, E.; Ross, RP.; Fitzgerald, GF.; Stanton, C. Rapid screening method for analyzing the conjugated linoleic acid production capabilities of bacterial cultures. Appl. Environ. Microbiol. 73, 7,23332337,(2007).
- Shantha, N.; Decker, EA.; Henning, B. Comparison of methylation methods for the quantification of Conjugated Linoleic Isomer. J. AOAC Int. 76,664649, (1993).
- Biasini, M.; Bienert, S.; Waterhouse, A.; Arnold, K.; Studer, G.; Schmidt, T. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information., Nucleic Acids Research. 42, W252, 8, (2014).
- Dassault Systèmes BIOVIA, Discovery Studio Modeling Environment. (2015).
- Hanwell, MD.; Curtis, DE.; Lonie, DC.; Vandermeerschd, T.; Zurek, E.; Hutchison, GR.; Avogadro. An advanced semantic chemical editor, visualization, and analysis platform. J. Cheminform. 4, 117, (2012).
- Morris, GM.; Huey, R.; Lindstrom, W.; Sanner, MF.; Belew, RK.; Goodsell, DS. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 30, 278591, (2009).
- Kim YJ, Liu RH, Bond DR, Russell JB. Effect of linoleic acid concentration on conjugated linoleic acid production by Butyrivibrio fibrisolvens A38. Appl. Environ. Microbiol. 66, 12, 52265230, (2000).
- van Nieuwenhove CP, Oliszewski R, Gonzalez SN, Perez Chaia AB.Conjugated linoleic acid conversion by dairy bacteria cultured in MRS broth and buffalo milk. Lett. Appl. Microbiol. 44, 5,467474, (2007).
- Ogawa J, Kishino S, Ando A, Sugimoto S, Mihara K, Shimizu S. Production of conjugated fatty acids by lactic acid bacteria. J. Biosci. Bioeng. 100, 4, 355364, (2005).
- Chung SH, Kim IH, Park HG, Kang HS, Yoon CS, Jeong HY, Choi NJ, Kwon EG, Kim YJ. Synthesis of conjugated linoleic acid by human-derived Bifido bacterium breve LMC 017: utilization as a functional starter culture for milk fermentation. J. Agric. Food Chem. 56,9, 33113316, (2008).
- Tung Y Lin, Chin-Wen Lin, Chien-Hsing Lee. Conjugated linoleic acid concentration as affected by lactic cultures and added linoleic acid Food Chemistry. 67, 1, 5, (1999).
- Alonso L, Cuesta EP, Gilliand SE. Production of free conjugated linoleic acid by Lactobacillus acidophilus and Lactobacillus casei of human intestinal origin. J. Dairy Sci. 86, 6, 19411946, (2003).