APPLICATION OF RESPONSE SURFACE MODELING FOR OPTIMIZATION AND DETERMINATION OF MALONDIALDIALDEHYDE BY VORTEX-ASSISTED DISPERSIVE LIQUID-LIQUID MICROEXTRACTION AND GC-FID
- Malondialdehyde,
- Vortex-assisted dispersive liquid-liquid microextraction,
- Response surface methodology,
- Human serum analysis,
- GC-FID
Abstract
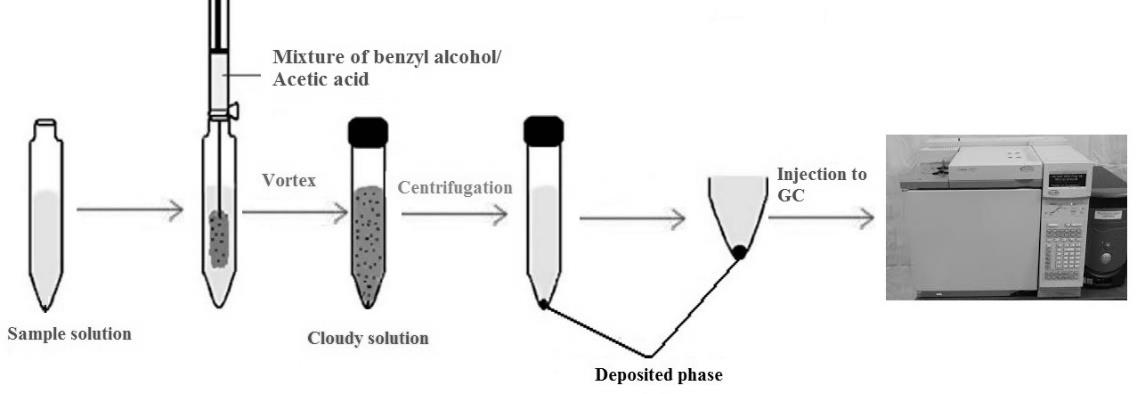
An analytical method based on vortex-assisted dispersive liquid-liquid microextraction and gas chromatography-flame ionization detection is presented for the extraction and determination of malondialdehyde )MDA( in blood plasma of human. Various parameters affecting the extraction efficiency such as type and volume of extraction and dispersive solvents, vortex and centrifuge times, volume, ionic strength and pH of the sample solution were evaluated using, one-variable-at-a-time and response surface methodology. In order to optimize the MDA extraction and determination, seven factors in five- levels were used for design of experiments (DOE). Under optimum extraction condition, this method showed linear range of calibration curve between 10–1150 μg L-1. The detection limit of the proposed method was found to be 0.8 μg L-1 with a relative standard deviation better than 5.5% (n=10) for blood serum samples. Enrichment factor was calculated to be 175 fold and the total analysis time including microextraction was about 13 min. The method was successfully applied for the analysis of MDA in blood plasma of human.
References
2. M. Rosenblat, R. Coleman, M. Aviram, Atherosclerosis 163, 17, (2002).
3. I. Delimaris, E. Faviou, G. Antonakos, E. Stathopoulou, A. Zachari, Clin. Biochem. 40, 1129, (2007).
4. A. M. Domijan, J. Ralić, S. Radić Brkanac, L. Rumora, T. Žanić‐Grubišić, Biomed. Chromatogr. 29, 41, (2015).
5. K. -C. Hsu, P. -F. Hsu, Y. -C. Chen, H. -C. Lin, C. -C. Hung, P. -C. Chen, Y. -L. Huang, J. Chromatogr. B 1019, 112, (2016).
6. E. M. Gioti, Y. C. Fiamegos, D. C. Skalkos, C. D. Stalikas, J. Chromatogr. A 1152, 150, (2007).
7. D. Tsikas, S. Rothmann, J. Y. Schneider, M. -T. Suchy, A. Trettin, D. Modun, N. Stuke, N. Maassen, J. C. Frölich, J. Chromatogr. B 1019, 95, (2015).
8. B. Liu, Y. Qi, M. Li, Y. Gao, X. Chen, Z. -T. Wang, Chin. Sci. Bull. 12, 36, (2010).
9. D. W. Wilson, H. N. Metz, L. M. Graver, P. S. Rao, Clin. Chem. 43, 1982, (1997).
10. S. H. Hashemi, M. Kaykhaii, F. Tabehzar, J. Iran Chem. Soc. 13, 733, (2016).
11. H. Hashemi, M. Khajeh, M. Kaykhaii, Anal. Methods 5, 2778, (2013).
12. M. Khajeh, M. Kaykhaii, M. Mirmoghaddam, H. Hashemi, J. Environ. Anal. Chem. 89, 981, (2009).
13. H. Uslu, D. Datta, D. Santos, H. S. Bamufleh, C. Bayat, Chem. Engineer. J. 299, 342, (2016).
14. M. Kaykhaii, A. Khatibi, J. Iran Chem. Soc. 8, 374, (2011).
15. M. Kaykhaii, H. Yahyavi, M. Hashemi, M. R. Khoshroo, 408, 4907, (2016).
16. J. -L. Chen, Y. -J. Huang, C. -H. Pan, C. -W. Hu, M. -R. Chao, Free Radic. Biol. Med. 51, 1823, (2011).
17. H. -S. Shin, J. Chromatogr. B 877, 3707, (2009).
18. K. Fujioka, T. Shibamoto, J. Agric. Food Chem. 53, 4708, (2005).
19. R. Heydari, S. Zarabi, Anal. Methods 6, 8469, (2014).
20. J. Gañán, D. Pérez-Quintanilla, S. Morante-Zarcero, I. Sierra, J. Hazard. Mater. 260, 609, (2013).
21. A. Mehdinia, A. Ghassempour, H. Rafati, R. Heydari, Anal. Chem. Acta 587, 82, (2007).
22. M. Rezaee, Y. Assadi, M. -R. M. Hosseini, E. Aghaee, F. Ahmadi, S. Berijani, J. Chromatogr. A 1116, 1, (2006).
23. S. H. Hashemi, M. Kaykhaii, R. Dehvari, Current Chromatogr. 4, 1, (2017).
24. M. Nassiri, M. Kaykhaii, S. H. Hashemi, M. Sepahi, Iran. J. Chem. Chem. Eng. 37, 89, (2018).
25. M. Khajeh, M. Kaykhaii, S. H. Hashemi, M. Shakeri, J. Food Compos. Anal. 33, 32, (2014).
26. J. Vichapong, R. Burakham, S. Srijaranai, Talanta 117, 221, (2013).
27. E. Yiantzi, E. Psillakis, K. Tyrovola, N. Kalogerakis, Talanta 80, 2057 (2010).
28. V. Hosseinpour, M. Kazemeini, A. Mohammadrezaee, Appl. Catal. A 394, 166 (2011).
29. K. Yetilmezsoy, S. Demirel, R. J. Vanderbei, J. Hazard. Mater. 171, 551, (2009).
30. M. Mirmoghaddam, M. Kaykhaii, M. Hashemi, Anal. Methods 8, 2456 (2016).
31. J. Lovrić, M. Mesić, M. Macan, M. Koprivanac, M. Kelava, V. Bradamante, Periodicum biologorum 110, 63, (2008).
32. A. H. S. Muñoz, M. P. Puga, K. Wrobel, M. E. G. Sevilla, K. Wrobel, Microchimica Acta 148, 285 (2004).